Abstract
Listeria monocytogenes is a food-borne pathogen that is able to form biofilms in food processing facilities. Biofilms are generally more resistant to antimicrobial agents, making it difficult to eradicate them during cleanup procedures. So far, little is known about the function of stress resistance mechanisms in biofilm formation and their resistance to disinfectants. In this study, we investigated the role of sigB, which encodes a major transcriptional regulator of stress response genes, in L. monocytogenes static and continuous-flow biofilm formation and its function in the resistance of biofilm cells to the disinfectants benzalkonium chloride and peracetic acid. Quantitative real-time PCR and promoter reporter studies showed that sigB is activated in static and continuous-flow biofilms. Biofilm formation studies using an in-frame sigB deletion mutant and complementation mutant showed that the presence of SigB is required to obtain wild-type levels of both static and continuous-flow biofilms. Finally, disinfection treatments of planktonically grown cells and cells dispersed from static and continuous-flow biofilms showed that SigB is involved in the resistance of both planktonic cells and biofilms to the disinfectants benzalkonium chloride and peracetic acid.
The food-borne pathogen Listeria monocytogenes is a Gram-positive facultative anaerobic rod and the causative agent of listeriosis, which is often manifested as meningitis, encephalitis, sepsis gastroenteritis, and spontaneous abortions (47). L. monocytogenes is of great public health concern because its incidence is increasing in many European countries (13). It is estimated that 99% of the listeriosis cases are caused by contaminated food products (26). Since L. monocytogenes is widely present in rural environments, it contaminates raw materials used by the food industry, thereby facilitating transmission to food processing facilities. In the food processing environment, L. monocytogenes is expected to survive by the formation of biofilms on food processing equipment, in drains, and in pipes and subsequently disperses to contaminate food products (31, 43).
A biofilm is defined as a community of microorganisms that is attached to a surface (29). So far, two distinct morphologies for L. monocytogenes biofilms have been identified. Biofilms formed under static conditions consist of small rod-shaped cells attached as single cells, microcolonies, or a homogeneous layer (8, 20, 35), while biofilms formed under continuous-flow conditions consist of ball-shaped microcolonies that are surrounded by a dense network of knitted chains composed of elongated cells (33). Recently, it was shown that formation of continuous-flow biofilms was dependent on the activation and presence of the SOS response factor YneA (44). Biofilms are generally more resistant to antimicrobial agents and disinfectants than planktonic cells. L. monocytogenes biofilms have also been shown to be more resistant to various disinfectants compared with planktonically grown cells (6, 16, 30, 34, 36, 41). Proposed mechanisms for the increased resistance of biofilms are the restricted penetration of the biofilm, the slow growth rate of organisms in the biofilm, and the induction of resistance mechanisms in the biofilm (14, 23, 24). Previously, a role for stress response sigma factors in the resistance of Pseudomonas aeruginosa biofilms to disinfectants has been shown (11). For L. monocytogenes, the stress response sigma factor SigB appeared not to be essential for attachment to stainless steel surfaces (39). So far, little is known on the activation and function of stress resistance mechanisms in L. monocytogenes biofilm formation and the resistance of biofilms to disinfectants.
In this study we investigated the role of sigB, which encodes the major stress response regulator, in L. monocytogenes static and continuous-flow biofilm formation and resistance of biofilm-grown cells to the disinfectants benzalkonium chloride and peracetic acid. The autoregulatory alternative sigma factor SigB is the regulator of the class II stress genes, which encode a group of proteins that play roles in response to various stress conditions (4, 5, 15, 45). SigB recognizes alternative −35 and −10 promoter sequences (GTTT-N13-17-GGGWAT) that are located in front of the class II stress genes (21). sigB is cotranscribed with seven regulatory genes from two operons, which are involved in posttranslational regulation of SigB (10, 19). Previously, it was shown in L. monocytogenes that the expression of a large number of genes and small rRNAs is regulated by SigB (1, 2, 18, 28). Furthermore, a role for SigB in the resistance of planktonic cells to benzalkonium chloride was recently identified (38). Investigating the role of this major stress response activator may provide clues to the requirements of stress genes in L. monocytogenes static and continuous-flow biofilm formation and disinfectant resistance.
MATERIALS AND METHODS
Strains, media, and plasmids.
L. monocytogenes strain EGD-e and its derivatives (Table 1) were grown in brain hearth infusion (BHI) broth (Becton Dickinson, Le Pont de Claix, France). A genomically expressed sigB promoter reporter strain was constructed using the site-specific integration plasmid pIMK-Pr.sigB-EGFP. This plasmid is a derivative of plasmid pIMK2-EGFP. The promoter region of sigB was amplified using primers sigB-1 and sigB-2 (Table 2) and cloned into pIMK2-EGFP as a SacI-NcoI fragment, which replaced the constitutively active Phelp promoter in front of enhanced green fluorescent protein (EGFP). The sigB genomic complementation mutant was constructed with the site-specific integration plasmid pIMK-sigB. This plasmid was made by replacing the EGFP of plasmid pIMK-Pr.sigB-EGFP with the sigB gene. sigB was amplified with primers sigB-3 and sigB-4 (Table 2) and cloned as an NcoI-SmaI fragment, thereby using the terminator present in plasmid pIMK-Pr.sigB-EGFP behind the EGFP gene.
TABLE 1.
Bacterial strains and plasmids used in this study
| L. monocytogenes strain or plasmid | Relevant genotype or characteristics | Reference or source |
|---|---|---|
| Strains | ||
| EGD-e | Wild-type serotype 1/2a strain | 17 |
| ΔsigB mutant | EGD-e ΔsigB | 9 |
| EGD-e:Pr.sigB-EGFP | EGD-e: genomic expression of EGFP from the sigB promoter using plasmid pIMK-Pr.sigB-EGFP | This study |
| sigB-c mutant | EGD-e ΔsigB: genomic complementation of sigB using plasmid pIMK-sigB | This study |
| Plasmids | ||
| pIMK2-EGFP | Kanr; pIMK2 derivative containing EGFP | 46 |
| pIMK-Pr.sigB-EGFP | Kanr; pIMK2-EGFP derivative containing the sigB promoter in front of EGFP | This study |
| pIMK-sigB | Kanr; pIMK2-EGFP derivative containing the sigB gene | This study |
TABLE 2.
PCR primers used in this study
| Primer | Sequence (5′-3′)a |
|---|---|
| sigB-1 | GTGGGAGCTCTTTAGAGAGCATTTACTGGTGA |
| sigB-2 | GTGGCCATGGCTTCACCCCATCTAATTTTAAG |
| sigB-3 | GCCCATGGAGGTGGAGGAGAATGCCA |
| sigB-4 | GCCCCGGGTGATTCAACTGCCTTGTTCAT |
| sigB-fwd | GCCGCTTACCAAGAAAATGG |
| sigB-rev | AATATTTTCGGGCGATGGAC |
| tpi-fwd | AACACGGCATGACACCAATC |
| tpi-rev | CACGGATTTGACCACGTACC |
| rpoB-fwd | CGTCGTCTTCGTTCTGTTGG |
| rpoB-rev | GTTCACGAACCACACGTTCC |
| 16S-rRNA-fwd | GATGCATAGCCGACCTGAGA |
| 16S-rRNA-rev | TGCTCCGTCAGACTTTCGTC |
Nucleotides introduced to create restriction sites are underlined.
Biofilm formation. (i) Static biofilm formation.
Static biofilms were grown in 12-well polystyrene microtiter plates (Greiner Bio-One, Frickenhausen, Germany). Overnight-grown cultures (18 h at 20°C) were inoculated (1%) in wells containing 3 ml BHI broth. The plates were incubated at 20°C for 48 h, and the medium was removed. The biofilms were washed three times with phosphate-buffered saline (PBS) (Merck, Darmstadt, Germany) and dispersed in 1 ml PBS by pipetting rigorously. Complete removal of the biofilms was verified by staining the wells with 0.1% crystal violet (Merck, Darmstadt, Germany). Serial dilutions were made in PBS, and appropriate dilutions were plated on BHI agar. The agar plates were incubated for 2 days at 30°C, and colonies were enumerated. Static biofilm formation was determined in two independent biological experiments using two replicates each.
(ii) Continuous-flow biofilm formation.
Continuous-flow biofilms were grown in a flow cell (BST FC 281; Biosurface Technologies Corporation, Bozeman, MT) at 20°C. Overnight-grown cultures (18 h at 20°C) were diluted in BHI (1%), and the flow chambers were inoculated. After 1-h bacterial adhesion, BHI medium was pumped through the flow cell with a flow of 10 ml/h. After 48 h, the biofilms were harvested and dispersed in 10 ml PBS. Cells were serial diluted in PBS, and appropriate dilutions were plated on BHI agar. Colonies were enumerated after 2 days of incubation at 30°C. Continuous-flow biofilms were quantified in two independent biological experiments using two replicates each.
Quantitative real-time PCR.
Cultures or biofilms were quenched in RNAprotect (Qiagen, Venlo, Netherlands) according to the manufacturer's protocol and harvested. RNA extraction was performed as described previously (46). Superscript III reverse transcriptase was used for synthesis of first-strand cDNA using 1 μg of total RNA. Absence of chromosomal DNA was verified for each RNA sample by omitting the Superscript III reverse transcriptase. Quantitative real-time PCRs were performed using the 2× Sybr green PCR Master Mix (Applied Biosystems, Nieuwerkerk a/d IJssel, Netherlands) and 200 nM primers in a total volume of 20 μl. The primers used for quantitative real-time PCR analyses were sigB-fwd and sigB-rev for sigB, tpi-fwd and tpi-rev for tpi, rpoB-fwd and rpoB-rev for rpoB, and 16S-rRNA-fwd and 16S-rRNA-rev for 16S rRNA (Table 2). Reactions were run on a ABI Prism 7000 sequence detection system (Applied Biosystems, Nieuwerkerk a/d IJssel, Netherlands) with an initial step of 10 min at 95°C and 40 cycles of 15 s at 95°C and 1 min at 60°C. A dissociation curve was added to verify single product formation. Furthermore, a calibration curve was generated for each primer set to calculate the efficiency of the PCRs. Expression levels of sigB were normalized using the average expression of the tpi, rpoB, and 16S rRNA housekeeping genes.
Microscopy.
Phase-contrast and fluorescence microscopy was performed using a BX41 microscope (Olympus, Zoeterwoude, Netherlands). Expression of EGFP was visualized using the MNIBA3 filter (Olympus, Zoeterwoude, Netherlands).
Disinfection treatments.
One milliliter of planktonic cells (24 h, 20°C) and cells dispersed from static and continuous-flow biofilms (log10 8.8 ± 0.3 CFU) were centrifuged for 2 min at 5,000 × g and resuspended in 1 ml sterile water. Benzalkonium chloride (Merck, Darmstadt, Germany) or peracetic acid (Sigma-Aldrich, Steinheim, Germany) was added in a final concentration of 20 μg/ml. Cells were exposed up to 15 min at 20°C, and samples were diluted (1:10) in neutralizing liquid (3 g/liter lecithin [VWR International Ltd, Poole, United Kingdom], 3% [vol/vol] Tween 80 [Merck, Hohenbrunn, Germany], 5 g/liter sodium thiosulfate [VWR International Ltd, Poole, United Kingdom], 1 g/liter l-histidine [Sigma-Aldrich, Steinheim, Germany], 0.34 g/liter potassium dihydrogen phosphate [Merck, Darmstadt, Germany]). Samples were serial diluted in PBS and plated on BHI agar. Colonies were enumerated after 3 to 5 days of incubation at 30°C. Disinfection treatments were performed in two independent biological replicates.
Data analyses.
Inactivation curves of the disinfectant treatments were fitted with the reparameterized Gompertz model (48) using the following equation:
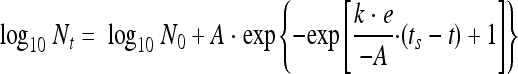 |
(1) |
where A is the difference between the surviving population and the initial population (log10 CFU/well), k is the maximum specific inactivation rate (log10/min), and ts is the duration of the shoulder (min). The inactivation curves were fitted in Microsoft Excel by minimizing the residual sum of squares using the Excel Solver add-in.
Significant differences in biofilm formation and sigB expression and between parameter estimates of the fitted inactivation curves were identified using the Student t test (P < 0.05).
RESULTS
SigB is activated during biofilm formation.
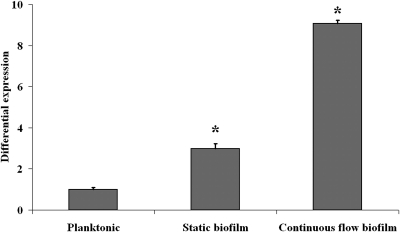
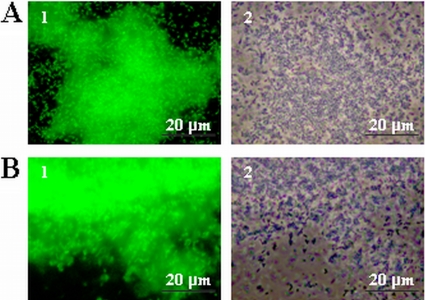
To investigate the possible activation of sigB during L. monocytogenes biofilm formation, the relative expression of sigB was analyzed with quantitative real-time PCR (Fig. 1). Compared with expression in planktonic cells, sigB expression was induced 3-fold in static biofilms and 9-fold in continuous-flow biofilms. The promoter activity of sigB in biofilms was further analyzed with a promoter reporter (Fig. 2). Expression of EGFP was not observed with planktonic cells (results not shown). Both static biofilm formation (Fig. 2A) and continuous-flow biofilm formation (Fig. 2B) resulted in high expression of EGFP, indicating that sigB is specifically activated during biofilm formation. Furthermore, expression of EGFP in continuous flow biofilms appeared to be more intense compared with expression in static biofilms, which corresponds with the results from the quantitative real-time PCR experiments.
FIG. 1.
Activation of sigB during biofilm formation. The graph shows differential expression of sigB between 48-h planktonic cultures, 48-h static biofilms, and 48-h continuous-flow biofilms grown at 20°C in BHI. Expression of sigB in planktonic cultures is set at 1. *, significantly different from planktonic cultures (P < 0.05, t test).
FIG. 2.
Activation of the sigB promoter during static (A) and continuous-flow (B) biofilm formation. Micrographs show fluorescence (1) and phase-contrast (2) pictures of cells expressing EGFP from the sigB promoter after 48 h of biofilm formation in BHI at 20°C.
SigB is important for biofilm formation.
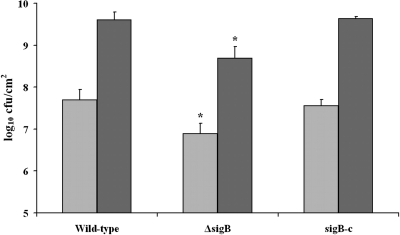
The impact of SigB on static and continuous-flow biofilm formation was assessed using the wild-type strain, an in-frame deletion mutant, and a complementation mutant (Fig. 3). No difference in planktonic growth between the wild-type and the ΔsigB mutant strain was observed (results not shown). The ΔsigB mutant showed a significant deficiency in biofilm formation under both static and continuous-flow conditions compared with the wild-type strain. This deficiency was restored in the complementation mutant, showing that SigB is an important factor during biofilm formation.
FIG. 3.
Comparative analysis of static and continuous-flow biofilm formation between the wild-type strain, the ΔsigB strain, and the complementation mutant (sigB-c). The graph shows biofilm formation in BHI at 20°C after 48 h under static (light gray) and continuous-flow (dark gray) conditions. *, significantly different from wild-type strain (P < 0.05, t test).
SigB is important for biofilm resistance against disinfection treatments.
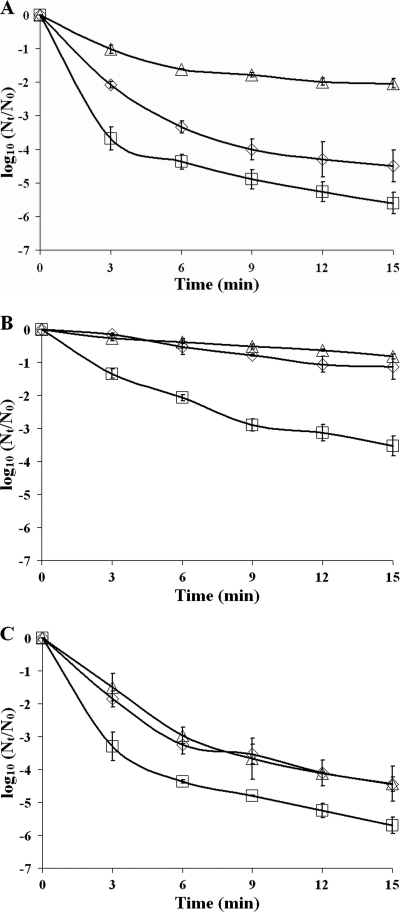
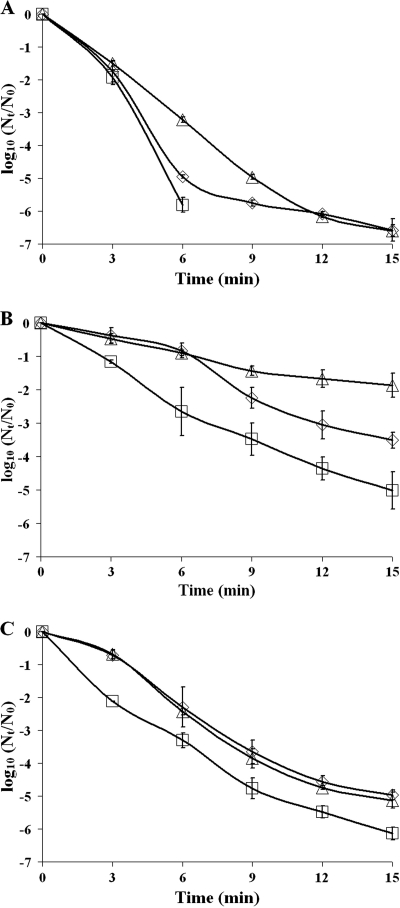
To investigate the role of SigB in the disinfection resistance of planktonic cells and biofilms, wild-type and mutant planktonic cells and cells dispersed from static and continuous-flow biofilms were exposed to the disinfectants benzalkonium chloride and peracetic acid. The inactivation curves were modeled, and parameter estimates were determined. Resistance against disinfection treatments is determined by the surviving population (A), the maximum specific inactivation rate (k), and the duration of the shoulder (ts). As reflected by the wild-type inactivation curves (Fig. 4) and all three of their parameter estimates (Table 3), dispersed static biofilm cells and not-dispersed continuous-flow biofilm cells showed increased resistance against benzalkonium chloride compared with planktonic grown cells. Furthermore, a role for SigB in the resistance of planktonic and biofilm cells against benzalkonium chloride was shown. Planktonic and dispersed static and continuous-flow biofilm cells of the ΔsigB mutant showed lower resistance to benzalkonium chloride treatments than wild-type cells, which is reflected by the lower surviving population (static and continuous-flow biofilm cells), the higher maximum inactivation rate (planktonic and static biofilm cells), or the shorter duration of the shoulder (static biofilm cells) of the ΔsigB mutant (Fig. 4 and Table 3). Complementation of the ΔsigB mutant restored or even overcompensated the benzalkonium chloride resistance level (Fig. 4 and Table 3), showing the requirement of SigB for the benzalkonium chloride resistance of both planktonic and biofilm cells.
FIG. 4.
Disinfection treatment of planktonic and biofilm cells with benzalkonium chloride. The graphs show the inactivation of 24-h planktonic cells (A), cells from a 48-h dispersed static biofilm (B), and cells from a 48-h dispersed continuous-flow biofilm (C) treated with 20 μg/ml benzalkonium chloride for 15 min at 20°C of wild-type (diamonds), ΔsigB (squares), and sigB-c (triangles) strains. Nt, number of viable cells at time t; N0, number of viable cells at time zero.
TABLE 3.
Parameter estimates of the inactivation curves after treatment with disinfectants
| Strain | Growth condition | Benzalkonium chloride |
Peracetic acid |
||||
|---|---|---|---|---|---|---|---|
| A | k | ts | A | k | ts | ||
| Wild-type | Planktonic | −4.39 ± 0.56 | 0.70 ± 0.03 | 0.25 ± 0.06 | −6.32 ± 0.18 | 1.20 ± 0.03 | 1.52 ± 0.20 |
| Static biofilm | −1.32 ± 0.52a | 0.12 ± 0.01a | 1.92 ± 0.21a | −4.04 ± 0.42a | 0.39 ± 0.01a | 3.47 ± 0.90 | |
| Continuous-flow biofilm | −4.23 ± 0.13 | 0.62 ± 0.10 | 0.20 ± 0.53 | −5.31 ± 0.12 | 0.57 ± 0.06a | 2.02 ± 0.47a | |
| ΔsigB mutant | Planktonic | −5.14 ± 0.26 | 1.35 ± 0.17b | 0.25 ± 0.03 | −7.20 ± 0.03b | 1.61 ± 0.08b | 1.80 ± 0.08 |
| Static biofilm | −3.54 ± 0.46b | 0.39 ± 0.01a,b | 0.08 ± 0.24b | −5.39 ± 0.12a,b | 0.47 ± 0.10a | 0.74 ± 0.01a,b | |
| Continuous-flow biofilm | −5.24 ± 0.30b | 1.13 ± 0.29 | 0.17 ± 0.15 | −6.43 ± 0.29b | 0.59 ± 0.02a | 0.07 ± 0.15a,b | |
| sigB-c mutant | Planktonic | −1.97 ± 0.08b | 0.35 ± 0.03b | 0.29 ± 0.41 | −7.17 ± 0.13b | 0.67 ± 0.02b | 1.09 ± 0.12 |
| Static biofilm | −1.03 ± 0.15a | 0.06 ± 0.00a,b | 0.06 ± 0.97 | −2.01 ± 0.51a,b | 0.19 ± 0.02a,b | 0.99 ± 1.02 | |
| Continuous-flow biofilm | −4.38 ± 0.52a | 0.56 ± 0.05a | 0.51 ± 0.30 | −5.44 ± 0.37a | 0.61 ± 0.06 | 2.08 ± 0.04a | |
Parameter estimate significantly different from planktonic cells (P < 0.05; t test).
Parameter estimate significantly different from the wild-type strain (P < 0.05; t test).
Notably, both dispersed static and continuous-flow biofilm cells of the wild-type strain showed higher resistance to peracetic acid treatments than planktonically grown cells (Fig. 5 and Table 3). Static biofilm cells showed a higher surviving population and a lower maximum inactivation rate than planktonically grown cells, while continuous-flow biofilm cells showed a lower maximum inactivation rate and a longer duration of the shoulder. Also, a role for SigB in the resistance of planktonic and dispersed biofilm cells against peracetic acid treatments was identified. Planktonically grown cells of the ΔsigB mutant showed a lower surviving population and a higher maximum inactivation rate than wild-type cells, and dispersed static and continuous-flow biofilm cells of the ΔsigB mutant showed a lower surviving population and a shorter duration of the shoulder than wild-type cells (Fig. 5 and Table 3). The role of SigB in the peracetic acid resistance of planktonic and biofilm cells was further emphasized by the fact that complementation of the ΔsigB mutant restored the resistance against peracetic acid of dispersed continuous-flow biofilm cells to wild-type levels and overcompensated the resistance of planktonic and dispersed static biofilm cells (Fig. 5 and Table 3), which resulted in increased resistance against peracetic acid.
FIG. 5.
Disinfection treatment of planktonic and biofilm cells with peracetic acid. The graphs show the inactivation of 24-h planktonic cells (A), cells from a 48-h dispersed static biofilm (B), and cells from a 48-h dispersed continuous-flow biofilm (C) treated with 20 μg/ml peracetic acid for 15 min at 20°C of wild-type (diamonds), ΔsigB (squares), and sigB-c (triangles) strains. Data points below the detection limit (log10 [Nt/N0] ≈ −6.5) are not shown in the graphs.
DISCUSSION
In this study we investigated the role of SigB in static and continuous-flow biofilm formation and the resistance of biofilm cells against the disinfectants benzalkonium chloride and peracetic acid. Our results show that the expression of sigB was specifically induced in static and continuous-flow biofilms. Furthermore, an in-frame deletion and complementation mutant showed that SigB is required to reach wild-type levels of both static and continuous-flow biofilms. It has been suggested in the past that the biofilm mode of growth results in microniches in which bacteria experience stress and therefore activate various stress resistance mechanisms (12). So far, not much is known of the activation and requirement of stress response genes in L. monocytogenes biofilm formation. One study has shown that SigB is not essential for the initial surface attachment of L. monocytogenes to stainless steel (39). To our knowledge, our study is the first that shows the involvement of SigB in L. monocytogenes biofilm formation. For Staphylococcus aureus and Staphylococcus epidermidis, a role for SigB in biofilm behavior has also been established previously. In these organisms, SigB-dependent biofilm-forming behavior involved regulation of the ica operon (22, 32). These studies indicate that, besides activation of SigB in biofilms due to local stresses encountered in these biofilms and thereby increasing the viability, in some bacteria certain stress signals can activate biofilm-forming behavior by activation of SigB and its regulon members.
We furthermore investigated the role of SigB in the resistance of static and continuous-flow biofilms to the disinfectants benzalkonium chloride and peracetic acid. Benzalkonium chloride and peracetic acid are commonly used disinfectants in the food industry (25). Commonly used concentrations of disinfectants in the food industry are 200 to 800 μg/ml for benzalkonium chloride and 0.5 to 5% for peracetic acid. However, it has been shown that L. monocytogenes is capable of surviving these disinfectants in, for instance, turkey processing plants and fish smokehouses (3, 27, 40), resulting in the development of resistance. In particular, L. monocytogenes biofilms have been shown to be capable of surviving disinfection treatments with benzalkonium chloride and peracetic acid (36, 41). So far, not much is known of the mechanisms behind the increased resistance of L. monocytogenes biofilms compared with planktonic cells. Benzalkonium chloride is thought to work by disruption and dissociation of the lipid bilayer of the cell membrane, resulting in leakage, while peracetic acid is expected to work as an oxidizing agent that produces hydroxyl radicals, which subsequently attack essential cell components (25). It has been shown previously that the resistance of L. monocytogenes planktonic cells to benzalkonium chloride is related to the fatty acid composition of the cell membrane and the induction of the efflux pump MdrL (37, 42). Furthermore, a role for the SigB response in benzalkonium chloride resistance has been established, since planktonic cells of a sigB deletion mutant showed enhanced sensitivity to benzalkonium chloride exposure (38). So far, no resistance mechanisms for peracetic acid have been shown for both planktonic and biofilm cells. In our study, increased resistance of dispersed static biofilm cells of the wild-type strain was observed after exposure to both benzalkonium chloride and peracetic acid compared with planktonically grown cells, while increased resistance of dispersed continuous-flow biofilm cells was shown only after peracetic acid treatments. A role for SigB in disinfectant resistance was shown with the reduced survival of the ΔsigB mutant in planktonic, static biofilm, and continuous-flow biofilm cells compared with that of the wild-type strain after exposure to both benzalkonium chloride and peracetic acid. However, the impact of SigB on the resistance of planktonic and biofilm cells against both disinfectants appeared to vary. A 3-fold induction of sigB expression was observed with static biofilm cells compared with planktonic cells, which is consistent with the observed increased resistance of these cells against both disinfectants. However, a 9-fold induction of sigB expression was observed with continuous-flow biofilm cells compared with planktonic cells, while increased resistance of continuous-flow biofilm cells was observed only after peracetic acid exposure and not after benzalkonium chloride exposure. Furthermore, continuous-flow and/or static biofilm cells of the wild-type strain showed increased resistance against benzalkonium chloride and/or peracetic acid exposure compared with planktonically grown cells of the ΔsigB mutant. These results indicate that SigB is not the only factor that is involved in the resistance of L. monocytogenes biofilms against disinfectants. Furthermore, induction of stress resistance genes, such as sigB, in biofilms probably indicates that bacteria already experience stress in these biofilms. Depending on the nature of this stress, bacteria might be adapted to resist specific types of disinfectants, while other types of disinfectants still kill these biofilm cells equally effectively. For Pseudomonas aeruginosa it has previously been shown that biofilms grown under continuous-flow conditions in a drip flow reactor experience DNA damage as a result of endogenous oxidative stress (7). For L. monocytogenes, it has previously been shown that oxidative stress results in the activation of sigB and that a ΔsigB mutant is less resistant to oxidative stress (4, 15). Exposure of continuous-flow biofilm cells to benzalkonium chloride, which targets the cell membrane, might result in a cumulative stress effect of DNA damage and cell membrane damage that is more difficult to resist. High expression of sigB in continuous-flow biofilm cells therefore might not be sufficient to increase the survival after benzalkonium chloride exposure. On the other hand, peracetic acid is an oxidizing agent that potentially targets DNA. High expression of sigB in collaboration with activated DNA repair mechanisms apparently increases the resistance of continuous-flow biofilm cells against this disinfectant. Previous studies have shown a number of SigB-regulated genes that could play a role in benzalkonium chloride or peracetic acid resistance, such as genes that encode proteins with a function in cell membrane biogenesis, lipid transport and metabolism, and oxidative stress resistance (18, 21).
In conclusion, our study highlighted the impact of SigB on L. monocytogenes biofilm formation and disinfection resistance. The SigB-regulated genes that contribute to the increased resistance of static and continuous-flow biofilm cells against disinfection treatments remain to be elucidated in future studies.
Footnotes
Published ahead of print on 1 October 2010.
REFERENCES
- 1.Abram, F., E. Starr, K. A. Karatzas, K. Matlawska-Wasowska, A. Boyd, M. Wiedmann, K. J. Boor, D. Connally, and C. P. O'Byrne. 2008. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74:6848-6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abram, F., W. L. Su, M. Wiedmann, K. J. Boor, P. Coote, C. Botting, K. A. Karatzas, and C. P. O'Byrne. 2008. Proteomic analyses of a Listeria monocytogenes mutant lacking σB identify new components of the σB regulon and highlight a role for σB in the utilization of glycerol. Appl. Environ. Microbiol. 74:594-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagge-Ravn, D., K. Gardshodn, L. Gram, and B. F. Vogel. 2003. Comparison of sodium hypochlorite-based foam and peroxyacetic acid-based fog sanitizing procedures in a salmon smokehouse: survival of the general microflora and Listeria monocytogenes. J. Food Prot. 66:592-598. [DOI] [PubMed] [Google Scholar]
- 4.Becker, L. A., M. S. Cetin, R. W. Hutkins, and A. K. Benson. 1998. Identification of the gene encoding the alternative sigma factor σB from Listeria monocytogenes and its role in osmotolerance. J. Bacteriol. 180:4547-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berrang, M. E., J. F. Frank, and R. J. Meinersmann. 2008. Effect of chemical sanitizers with and without ultrasonication on Listeria monocytogenes as a biofilm within polyvinyl chloride drain pipes. J. Food Prot. 71:66-69. [DOI] [PubMed] [Google Scholar]
- 7.Boles, B. R., and P. K. Singh. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc. Natl. Acad. Sci. U. S. A. 105:12503-12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chae, M. S., H. Schraft, L. Truelstrup Hansen, and R. Mackereth. 2006. Effects of physicochemical surface characteristics of Listeria monocytogenes strains on attachment to glass. Food Microbiol. 23:250-259. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cochran, W. L., S. J. Suh, G. A. McFeters, and P. S. Stewart. 2000. Role of RpoS and AlgT in Pseudomonas aeruginosa biofilm resistance to hydrogen peroxide and monochloramine. J. Appl. Microbiol. 88:546-553. [DOI] [PubMed] [Google Scholar]
- 12.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 13.Denny, J., and J. McLauchlin. 2008. Human Listeria monocytogenes infections in Europe—an opportunity for improved European surveillance. Euro Surveill. 13:pii=8082. [PubMed] [Google Scholar]
- 14.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira, A., C. P. O'Byrne, and K. J. Boor. 2001. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl. Environ. Microbiol. 67:4454-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folsom, J. P., and J. F. Frank. 2006. Chlorine resistance of Listeria monocytogenes biofilms and relationship to subtype, cell density, and planktonic cell chlorine resistance. J. Food Prot. 69:1292-1296. [DOI] [PubMed] [Google Scholar]
- 17.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 18.Hain, T., H. Hossain, S. S. Chatterjee, S. Machata, U. Volk, S. Wagner, B. Brors, S. Haas, C. T. Kuenne, A. Billion, S. Otten, J. Pane-Farre, S. Engelmann, and T. Chakraborty. 2008. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e sigmaB regulon. BMC Microbiol. 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helmann, J. D. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2:135-141. [DOI] [PubMed] [Google Scholar]
- 20.Kalmokoff, M. L., J. W. Austin, X. D. Wan, G. Sanders, S. Banerjee, and J. M. Farber. 2001. Adsorption, attachment and biofilm formation among isolates of Listeria monocytogenes using model conditions. J. Appl. Microbiol. 91:725-734. [DOI] [PubMed] [Google Scholar]
- 21.Kazmierczak, M. J., S. C. Mithoe, K. J. Boor, and M. Wiedmann. 2003. Listeria monocytogenes sigma B regulates stress response and virulence functions. J. Bacteriol. 185:5722-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knobloch, J. K., S. Jager, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 72:3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis, K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45:999-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 25.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullapudi, S., R. M. Siletzky, and S. Kathariou. 2008. Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 74:1464-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliver, H. F., R. H. Orsi, L. Ponnala, U. Keich, W. Wang, Q. Sun, S. W. Cartinhour, M. J. Filiatrault, M. Wiedmann, and K. J. Boor. 2009. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics 10:641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 30.Pan, Y., F. Breidt, Jr., and S. Kathariou. 2006. Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 72:7711-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pritchard, T. J., K. J. Flanders, and C. W. Donnelly. 1995. Comparison of the incidence of Listeria on equipment versus environmental sites within dairy processing plants. Int. J. Food Microbiol. 26:375-384. [DOI] [PubMed] [Google Scholar]
- 32.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rieu, A., R. Briandet, O. Habimana, D. Garmyn, J. Guzzo, and P. Piveteau. 2008. Listeria monocytogenes EGD-e biofilms: no mushrooms but a network of knitted chains. Appl. Environ. Microbiol. 74:4491-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins, J. B., C. W. Fisher, A. G. Moltz, and S. E. Martin. 2005. Elimination of Listeria monocytogenes biofilms by ozone, chlorine, and hydrogen peroxide. J. Food Prot. 68:494-498. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez, A., W. R. Autio, and L. A. McLandsborough. 2008. Effect of surface roughness and stainless steel finish on Listeria monocytogenes attachment and biofilm formation. J. Food Prot. 71:170-175. [DOI] [PubMed] [Google Scholar]
- 36.Romanova, N. A., P. V. Gawande, L. Y. Brovko, and M. W. Griffiths. 2007. Rapid methods to assess sanitizing efficacy of benzalkonium chloride to Listeria monocytogenes biofilms. J. Microbiol. Methods 71:231-237. [DOI] [PubMed] [Google Scholar]
- 37.Romanova, N. A., P. F. Wolffs, L. Y. Brovko, and M. W. Griffiths. 2006. Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl. Environ. Microbiol. 72:3498-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan, E. M., C. G. Gahan, and C. Hill. 2008. A significant role for sigma B in the detergent stress response of Listeria monocytogenes. Lett. Appl. Microbiol. 46:148-154. [DOI] [PubMed] [Google Scholar]
- 39.Schwab, U., Y. Hu, M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factor sigmaB is not essential for Listeria monocytogenes surface attachment. J. Food Prot. 68:311-317. [DOI] [PubMed] [Google Scholar]
- 40.Soumet, C., C. Ragimbeau, and P. Maris. 2005. Screening of benzalkonium chloride resistance in Listeria monocytogenes strains isolated during cold smoked fish production. Lett. Appl. Microbiol. 41:291-296. [DOI] [PubMed] [Google Scholar]
- 41.Stopforth, J. D., J. Samelis, J. N. Sofos, P. A. Kendall, and G. C. Smith. 2002. Biofilm formation by acid-adapted and nonadapted Listeria monocytogenes in fresh beef decontamination washings and its subsequent inactivation with sanitizers. J. Food Prot. 65:1717-1727. [DOI] [PubMed] [Google Scholar]
- 42.To, M. S., S. Favrin, N. Romanova, and M. W. Griffiths. 2002. Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl. Environ. Microbiol. 68:5258-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tompkin, R. B. 2002. Control of Listeria monocytogenes in the food-processing environment. J. Food Prot. 65:709-725. [DOI] [PubMed] [Google Scholar]
- 44.van der Veen, S., and T. Abee. 2010. Dependence of continuous-flow biofilm formation by Listeria monocytogenes EGD-e on SOS response factor YneA. Appl. Environ. Microbiol. 76:1992-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Veen, S., T. Hain, J. A. Wouters, H. Hossain, W. M. de Vos, T. Abee, T. Chakraborty, and M. H. Wells-Bennik. 2007. The heat-shock response of Listeria monocytogenes comprises genes involved in heat shock, cell division, cell wall synthesis, and the SOS response. Microbiology 153:3593-3607. [DOI] [PubMed] [Google Scholar]
- 46.van der Veen, S., S. van Schalkwijk, D. Molenaar, W. M. de Vos, T. Abee, and M. H. Wells-Bennik. 2010. The SOS response of Listeria monocytogenes is involved in stress resistance and mutagenesis. Microbiology 156:374-384. [DOI] [PubMed] [Google Scholar]
- 47.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. van 't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]