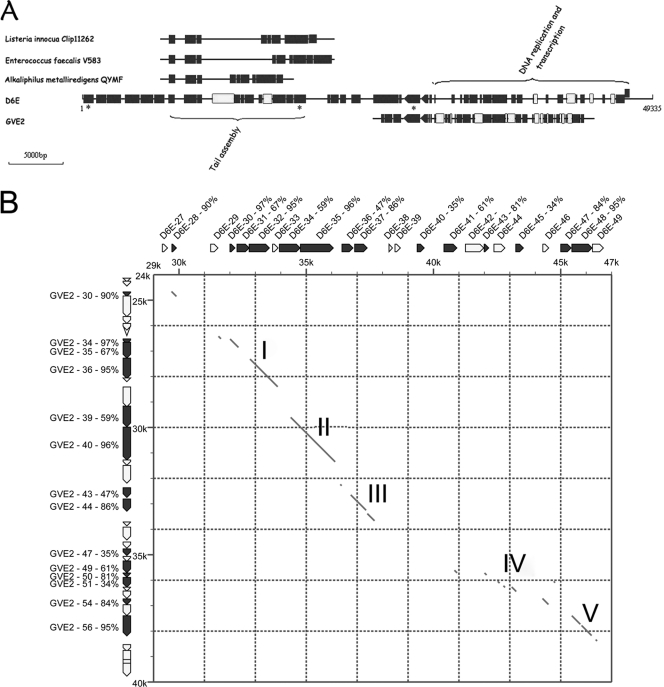
FIG. 3.
Mosaicism in bacteriophage D6E. Multiple alignments were generated with DNAMAN program (Lynnon Biosoft) using the ClustalW algorithm. (A) Comparative genomics among D6E, three mesophilic bacteriophages (Listeria innocua Clip 11262, Enterococcus faecalis V583, and Alkaliphilus metalliredigens QYMF), and deep-sea thermophilic bacteriophage GVE2. The full-length D6E genome is displayed, while the partial genomes of three mesophilic phages and GVE2 with similarities to D6E are shown. Black boxes represent genes with high similarities, and white boxes show genes with low similarities. (B) Comparison of DNA replication and transcription gene cluster between D6E and GVE2. The regions of D6E and GVE2 were compared by a dot-matrix-dot method. Gene positions are indicated in thousands. The diagonal lines show regions where the nucleotide sequences were matched. The nucleotide and protein sequence similarity extended five segments over the DNA transcription and replication region I∼V. The percentages show amino acid sequence similarities between ORFs from bacteriophages D6E and GVE2.