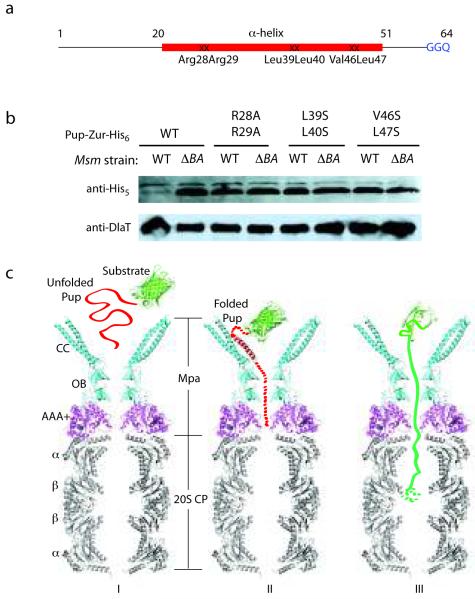
Figure 3.
Essentiality of the Pup helical region for proteasomal degradation supports a binding-induced folding recognition mechanism by Mpa. (a) Site-directed mutations in Pup resulted in abrogated degradation of a Pup-linear fusion by the Msm proteasome. The binding-induced helical region is in red. Mutated residues are indicated. (b) Equivalent cell numbers were analyzed from stationary phase cultures of wild type (WT) or proteasome-deleted (ΔprcBA, ΔBA) M. smegmatis synthesizing WT or mutated Pup-Zur-His6. Detection of the linear fusion proteins was done with anti-His5. DlaT (dihydrolipoamide acyltransferase) was the loading control. (c) Model for the targeting of pupylated proteins for degradation by Mpa and the mycobacterial proteasome. The Pup:Mpa1-234 complex structure (red and cyan) was placed over the homologous PAN AAA+ domain structure (PDB ID 3H4M, magenta), which was further overlaid on the Mtb proteasome core structure (PDB ID 2FHH, gray). Only a vertical central slice of the complex structure is shown for clarity. Pup is in red, and a model substrate (GFP) in green. See main text for details.