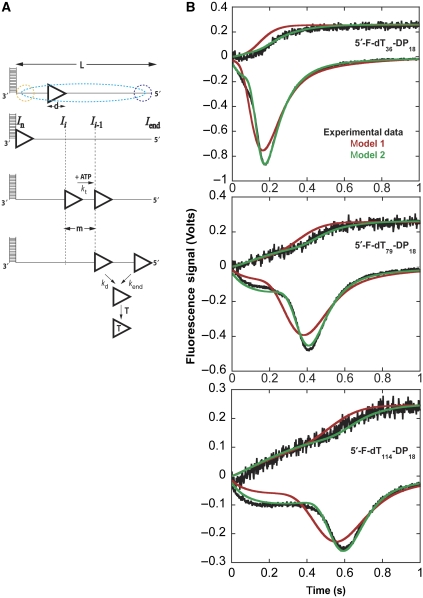
Figure 3.
Analysis of time courses of UvrD monomer translocation along 5′-ss/dsDNA using a sequential n-step model accounting for a non-random initial binding distribution. (A) Kinetic model for UvrD monomer translocation on 5′-ss/dsDNA as described in the text. UvrD monomers (triangle), high-specificity binding site (junction, yellow circle) and low-specificity binding sites (internal ssDNA sites and 5′-end, blue circle). UvrD is prevented from rebinding the DNA by binding to heparin (protein trap, T). Intermediate positions of UvrD along the ssDNA are shown as Ii, where In represents UvrD bound to the ss/dsDNA junction. (B) Global NLLS analysis of time courses monitoring dissociation of UvrD during translocation and arrival of UvrD at the 5′-end of 5′-F-dTL-DP18 (L=36, 79 and 114) as described in Supplementary data. A subset of the lengths analysed are shown, the rest are shown in Supplementary Figure S8. The coloured lines are simulations of the best-fit parameters (Supplementary Table S1) examining translocation models 1 and 2. The translocation models are summarized in the text and described further in Supplementary data.