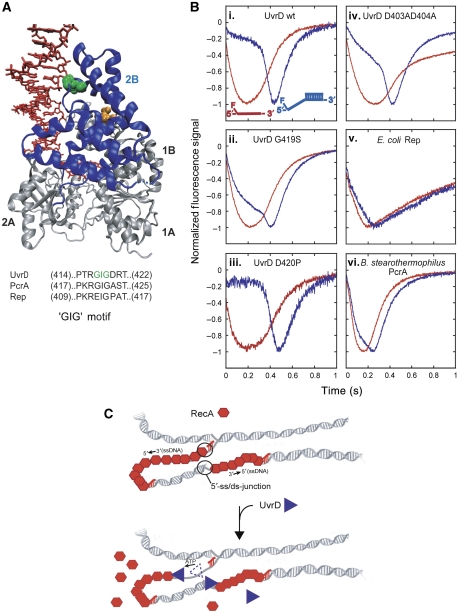
Figure 7.
‘GIG' motif of UvrD 2B domain is important for UvrD specificity for binding 5′-ss/dsDNA junctions. (A) Crystal structure of UvrD bound to a partial duplex with a 3′-ssDNA tail (Lee and Yang, 2006). The green space fill is the GIG (419) of the ‘GIG' motif in the 2B domain (blue ribbon). The orange space fills are residues D403 and D404. The sequence alignment of ‘GIG' motif in UvrD subfamily of SF1 helicase/translocases is shown. (B) Translocation time courses monitoring the arrival UvrD 2B domain mutants and other SF1 helicase/translocases at the 5′-end of ssDNA and 5′-ss/dsDNA substrates. The ssDNA (dT79, red) or 5′-ss/dsDNA (5′-dT79-DP18, blue) is labelled at the 5′-ssDNA end with fluorescein. The time courses are normalized to the peak signal change. (i–iv) wt-UvrD or UvrD 2B domain mutant translocation. (v) E. coli Rep translocation. (vi) B. stearothermophilus PcrA translocation (PcrA (40 nM), 20 mM Tris–HCl pH 7.5, 100 nM DNA, 10 mM NaCl, 10% (v/v) glycerol, 100 nM DNA, 2.5 mM ATP, 3 mM MgCl2 and 4 mg/ml heparin). (C) Proposal for physiological role of UvrD specificity for 5′-ss/dsDNA junctions. UvrD specifically targets RecA filaments that form on the lagging strand of collapsed replication forks. 5′-ss/dsDNA junctions provide a high-affinity site for UvrD to bind and invade the RecA filament, resulting in displacement of RecA by UvrD translocation along the ssDNA.