Abstract
Recent genome-wide studies have revealed a remarkable correspondence between nucleosome positions and exon-intron boundaries, and several studies have implicated specific histone modifications in regulating alternative splicing. In addition, recent progress in cracking the ‘splicing code’ shows that sequence motifs carried on the nascent RNA molecule itself are sufficient to accurately predict tissue-specific alternative splicing patterns. Together, these studies shed light on the complex interplay between RNA sequence, DNA sequence, and chromatin properties in regulating splicing.
Introduction and context
The fact that the majority of pre-mRNAs are spliced while they are still being transcribed [1] has led to the proposal that local chromatin structure and histone modifications may play direct roles in regulating splicing. Indeed, several studies have shown that specific histone modifications can affect the association of splicing factors with chromatin and the efficiency of the splicing process [2-4]. A link between nucleosome positioning and exon-intron boundaries was first proposed in 1991, long before the functional link between splicing and transcription was established [5]. Recent advances in the computational prediction and experimental verification of nucleosome positioning, based on underlying DNA sequence features [6-8], in combination with recently published high-resolution genome-wide maps of nucleosome positions [9,10] and histone modifications [11,12] in several organisms, have now enabled genome-wide comparisons of nucleosome positioning, histone modifications, and intron-exon architecture, revealing surprising correlations between these features and throwing light on the question of how chromatin features may help the splicing machinery to distinguish between exons and introns.
A distinct question is that of alternative splicing. Alternative splicing can generate many different transcripts from a single gene. Recent high-throughput transcriptome analyses have detected alternative splicing in approximately 95% of multi-exon human genes [13-15] and also extensively in mice [16], plants [17], flies [18,19], and yeast [20,21]. Alternative splicing is often regulated by trans-acting factors that are differentially expressed in different tissues or metabolic states and bind specific sequence or structural motifs on the pre-mRNA, resulting in alternative splicing [22-24]. It has long been a goal of the splicing field to crack the ‘splicing code’: to identify the pre-mRNA sequence features that can explain and predict not only the exact sites of constitutive splicing but also the features that determine tissue-specific alternative splicing patterns [25].
Previous work has successfully identified pre-mRNA-encoded features that define the precise boundaries of certain classes of constitutively spliced exons [26-28] and additional splicing enhancers and silencers [28,29], some of which have been shown to correlate with exon inclusion levels in specific tissues [16,28]. However, it has been suggested that pre-mRNA-encoded information alone is not sufficient to explain the recognition of short exons in a desert of long introns or the modulation of this process in alternative splicing [30]. The idea that local chromatin structure may add an additional regulatory layer to splicing, particularly to alternative splicing, is currently gaining momentum, and both correlative evidence and functional evidence are emerging [30-35]. Additional insights into the role of RNA sequence come from the recent demonstration that by taking hundreds of RNA features into account, a ‘splicing code’ based on pre-mRNA features is able to qualitatively predict tissue-specific alternative splicing for thousands of vertebrate exons [36]. Here, recent advances and current ideas on how chromatin and pre-mRNA sequence contribute to constitutive and alternative splicing are reviewed.
Major recent advances
In 2009 and 2010, several groups [37-42] used bioinformatic approaches to uncover patterns from published genome-wide maps of nucleosome positioning in human T cells and in Caenorhabditis elegans, derived from deep sequencing of micrococcal nuclease-digested chromatin [9,10]. These recent bioinformatic analyses [37-42] now show that nucleosomes sit preferentially on exon sequences whereas introns are relatively depleted of nucleosomes (Figure 1). This remarkable arrangement was observed in several metazoan species and was found to be independent of transcriptional activity, suggesting that the arrangement is an inherent property of chromatin [37-39,41]. Strikingly, the average length observed for metazoan exons (140-150 base pairs) corresponds neatly with the length of DNA required to wrap a single nucleosome (147 base pairs) [39,41]. Further correlations to the splicing process were observed by several authors (e.g., exons flanked by weak splice sites or by long introns have a higher tendency to be bound by nucleosomes) [40,41], raising the idea that nucleosomes may help the splicing machinery by ‘marking’ exons that may otherwise be difficult to recognize (reviewed in [30-32]).
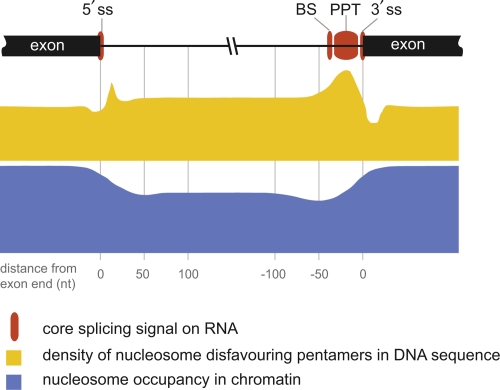
Figure 1. RNA, DNA, and chromatin features at exon/intron boundaries.
The positions of core RNA splicing signals (red boxes), DNA sequence properties (yellow profile), and nucleosome occupancy (blue profile) on human exon/intron boundaries are drawn approximately to scale according to the distance from the exon end. Core RNA splicing signals include the 5′ splice site (ss) (position -2 to +4), which has a variable sequence in higher eukaryotes and a human consensus of AGGUAAG. The branch site (BS) has a variable position at -20 to -30, directly 5′ of the polypyrimidine tract (PPT), and a variable sequence, with a human consensus of CUAAC. The PPT (position -5 to -20 or -30) is variable in length (in humans typically 10-25 pyrimidines) and strongly favors uracil. The 3′ ss (position -3 to +1) has a variable sequence, and a human consensus of CAGG [27]. Each of the core splice signals on the pre-mRNA is recognized specifically by a protein or ribonucleoprotein component of the splicing machinery [25-27]. Not shown are the numerous additional splicing regulatory elements, which reside in introns and exons and add specificity to constitutive and tissue-specific splicing [23,25,36]. Nucleosome occupancy (blue profile): The average relative nucleosome occupancy across the exon/intron junctions is shown to scale for human activated T cells (adapted from [39,40]). The vertical scale represents approximately 1.5-fold higher enrichment on exons compared with introns. Note the more profound depletion at the 3′ intron end. The extent of depletion at this site was found to correlate positively with the strength of the PPT [39]. DNA sequence properties (yellow profile): The density of pentamer sequences that disfavor nucleosome binding [6] was calculated for constitutive exon/intron junctions in [39] and is drawn approximately to scale. The maximum value on the vertical scale represents approximately 70% of sequences containing a nucleosome-disfavoring pentamer, and the minimum is approximately 40%. Guanine and cytosine (GC) content (not shown) may also play a role in nucleosome positioning. Exons have a higher GC content than surrounding intron regions, and this has been proposed to contribute to the observed increased nucleosome occupancy over exons relative to introns [31]. However, there is some disagreement on this point in the literature (for a discussion, see [32]).
By analysis of chromatin immunoprecipitation (ChIP) combined with high-throughput sequencing (ChIP-seq) and ChIP followed by microarray analysis (ChIP-chip) data sets [9,11,12], several studies have examined correlations between histone modifications and exon-intron boundaries and have reported conflicting results. Several authors conclude that specific histone modifications are enriched on exons compared with introns, suggesting an active marking mechanism [12,37,40,43], whereas others argue that these apparent enrichments are due mostly to nucleosome positioning, which is independent of modification status [38,39,41]. It has been suggested that these discrepancies may be due mainly to difficulties of normalization of ChIP data for nucleosome occupancy, compounded by the fact that occupancy studies and modification studies were performed with different techniques (micrococcal nuclease digestion versus ChIP) [30]. It is also possible that gene-specific differences exist but go undetected in global analyses.
To what extent can we understand the preference of nucleosomes for exonic sequences in terms of known pre-mRNA splicing signals? Figure 1 summarizes the RNA, DNA, and chromatin features at exon-intron boundaries. The preference of nucleosomes for exons appears to be a consequence not only of the higher guanine and cytosine content of exons [38,39,41] but also of a high density of sequences that repel nucleosomes [6] exactly at the intron-exon boundary and a depletion of these sequences within the exon itself relative to the adjacent intron [39] (Figure 1). Particularly interesting is the polypyrimidine tract (PPT), typically a long (10-20 nucleotides) run of uracil bases at the 3′ intron end in the pre-mRNA, that is specifically recognized by spliceosome components (Figure 1). At the DNA level, the PPT corresponds to poly T, one of the strongest nucleosome-repelling sequences identified by Kaplan et al. [6]. Thus, it is clear that splicing signal sequences play a role both at the RNA level, for recognition by RNA-binding proteins of the splicing machinery, and at the DNA level, in determining the positions of nucleosomes. An additional role of splice site sequences in increasing RNA flexibility has been proposed [42]. Is nucleosome positioning thus merely a coincidence of the RNA sequence or does it have a causal role in splicing? Experimental evidence beyond correlation is currently lacking but several possible roles have been proposed (summarized in Figure 2).
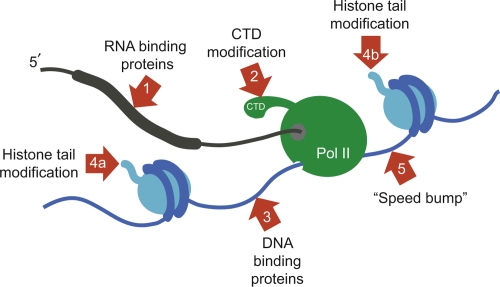
Figure 2. Integration of RNA, DNA, and chromatin signals in cotranscriptional regulation of splicing.
The nascent pre-mRNA molecule is shown in black, and the exon is shown as a thicker line. The large subunit of RNA polymerase II (Pol II) is shown in green, transcribing from left to right. The C-terminal domain (CTD) of Pol II is indicated. DNA is shown in blue, with exon-encoding sequences (thicker lines) wrapped around nucleosomes (light blue). Red arrows show different points at which the splicing machinery has been shown or proposed to operate. (1) RNA sequences (shown in Figure 1) are bound directly by the splicing machinery via protein-RNA interactions or ribonucleoprotein-RNA interactions [25]. Core signals shown in Figure 1 operate on all exons, whereas additional tissue-specific signals act in conjunction with tissue-specific splicing factors to ensure alternative splicing [23,36]. (2) The CTD is phosphorylated at different residues upon initiation, elongation, and termination and serves as a binding platform for other proteins. The CTD may affect pre-mRNA processing directly by recruiting splicing factors or indirectly by recruiting nucleosome remodelers and histone modifiers [47]. (3) When Pol II transcribes through a nucleosome, core histones are transferred behind the transcribing polymerase via a transient DNA loop [44]. Unassembled DNA or DNA in linker sequences presents an opportunity for splicing regulators to bind directly to DNA sequences. (4a, 4b) Histone tail modifications can recruit regulators of alternative splicing to chromatin [34,35]. These may be on reassembled nucleosomes behind Pol II (4a) or on nucleosomes in front of Pol II (4b). (5) Nucleosomes positioned on exons have been proposed to act as ‘speed bumps’, slowing down the polymerase immediately as it begins transcribing the exon upon which the nucleosome is positioned [30,39]. Such a mechanism may affect any of steps 1 to 4 by modulating the time available for interactions of trans-acting factors with the exon (at the DNA, RNA, or chromatin level) emerging behind the polymerase.
The models proposed so far fall into one of two non-mutually exclusive classes - the ‘recruitment’ models and the ‘kinetic coupling’ models [32] - and are summarized in Figure 2. Recruitment models favor the idea that nucleosome positioning or nucleosome modifications on exons guide the splicing machinery to the right place (Figure 2), whereas kinetic coupling proposes that the presence of a nucleosome in the path of the polymerase decreases the speed of transcription and thus allows more time for splicing to occur (Figure 2, arrow 5). In favor of such ‘speed bump’ models [30], single-molecule in vitro experiments have shown that the speed of RNA polymerase II (Pol II) transcription is modulated by nucleosomal barriers [44]. On the other hand, a recent in vivo study of Pol II transcription rates measured similar speeds on exonic and intronic sequences and showed that although splicing does indeed occur cotranscriptionally, it lags substantially behind the transcription process, being approximately twice as slow [45]. This raises the question of how much the chromatin features ahead of the polymerase can influence splicing events that take place far behind it (Figure 2). However, this study was limited to a small number of genes, and it is not known whether they contain exons that are regulated in a chromatin or Pol II elongation-dependent manner (or both).
If nucleosome positions do affect splicing, then how could such a system permit the vast amount of flexibility observed in alternative splicing? Several authors have proposed that nucleosome remodelers, a relaxation of nucleosome positioning at alternatively spliced exons, or tissue-specific histone modifications may account for tissue-specific differences in nucleosome positioning or properties and thus facilitate alternative splicing [30-32,39]. In support of this idea, ectopic recruitment of heterochromatin modifications to the vicinity of an alternative exon was found to reduce Pol II speed and to affect the splicing of that exon [34]. Furthermore, investigation of the fibroblast growth factor receptor 2 (FGFR2) locus revealed cell line-specific enrichment of histone H3 lysine 36 trimethylation (H3K36me3), which was shown to be required to promote the exclusion of one exon of the gene [35]. An adaptor protein recruits the proteins required for specific exclusion of this exon to the sites of H3K36 methylation, demonstrating a direct mechanistic link between cell type-specific chromatin modification and exon exclusion. Interestingly, the H3K36me3 enrichments were broadly spread across the FGFR2 locus and were not limited to the excluded exon, suggesting that nucleosome positioning does not play the major role here; it is the H3K36me3 modification that serves to recruit the necessary factors to the general vicinity of an alternatively spliced exon in the appropriate cell type, although sequence features of the pre-mRNA itself determine which exon is to be excluded.
Although it remains unclear to what extent alternative nucleosome positions play a role in alternative splicing, a recent study [36] demonstrates that pre-mRNA sequence features are an essential component and perhaps contain sufficient information for many alternative splicing events. The authors examined the splicing patterns of over 3000 alternative exons in 27 mouse tissues and extracted over 1000 RNA sequence features, including known and novel motifs and structural features. From this the authors compiled 200 features that were diagnostic and qualitatively predictive for tissue-specific alternative splicing. The predictive power of the code was tested by cross-validation and comparison with experimental data. Depending on the exon type, the code was able to correctly predict alternative splicing in central nervous system and muscle for 65-95% of test exons. Importantly, the code is combinatorial, and the authors conclude that large numbers of sequence features are required to ensure tissue-specific splicing. Although this study focuses on the idea that these are pre-mRNA sequence features, it is also possible that several of the newly identified features may work at the DNA level or at both RNA and DNA levels as is the case for the PPT [39] (Figure 1).
Future directions
In the future, it will be essential to determine the relative contributions of RNA sequence features, nucleosome positioning, and histone modifications to constitutive and alternative splicing. The data so far correlating nucleosome positioning to exon-intron architecture have been limited to few cell types. It will be important to see whether nucleosome positions are indeed different in different tissues, as has been proposed via thermodynamic competition with tissue-specific transcription factors [7] and observed for regulatory regions of specific loci [46]. If this is also the case on a genome-wide scale, then do alternative nucleosome positions correlate with alternatively spliced exons?
Furthermore, it would be extremely interesting to investigate whether any of the RNA sequence features that are predictive for alternative splicing [36] could play an additional role at the DNA level (e.g., as nucleosome positioning or repelling sequences [6] or as binding sites for site-specific DNA binding proteins that may compete with nucleosomes in a tissue-specific manner [7]).
Finally, it will be essential to go beyond global correlations and to determine cause and effect for specific loci. To unravel the relative contributions of RNA sequence, DNA sequence, and chromatin architecture, it will be essential to overcome the inherent difficulty in such experiments (i.e., that any change in DNA sequence changes the RNA sequence). Thus, it will be essential to devise strategies by which nucleosome positions can be modulated without affecting the underlying DNA sequence (e.g., by manipulating levels of remodelers or competing transcription factors).
Acknowledgments
Work in the author's laboratory is supported by the Austrian Academy of Sciences, the FWF (Fonds zur Förderung der wissenschaftlichen Forschung [Austrian Scientific Research Fund]), Gen-AU (Genomforschung in Österriech [Genome Research in Austria]), The Epigenome Network of Excellence (EU- FP6), and the FCT (Fundação para a Ciência e Tecnologia [Portugese Foundation for Science and Technology]).
Abbreviations
- ChIP
chromatin immunoprecipitation
- ChIP-chip
chromatin immunoprecipitation followed by microarray analysis
- ChIP-seq
chromatin immunoprecipitation followed by high throughput sequencing
- FGFR2
fibroblast growth factor receptor 2
- H3K36me3
histone 3 lysine 36 trimethylation
- Pol II
RNA polymerase II
- PPT
polypyrimidine tract
Competing Interests
The author declares that she has no competing interests.
The electronic version of this article is the complete one and can be found at: http://f1000.com/reports/b/2/74
References
- 1.Neugebauer KM. On the importance of being co-transcriptional. J Cell Sci. 2002;115:3865–71. doi: 10.1242/jcs.00073. [DOI] [PubMed] [Google Scholar]
- 2.Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell. 2009;33:450–61. doi: 10.1016/j.molcel.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allemand E, Batsche E, Muchardt C. Splicing, transcription, and chromatin: a ménage à trois. Curr Opin Genet Dev. 2008;18:145–51. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Sims RJ, 3rd, Millhouse S, Chen CF, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckmann JS, Trifonov EN. Splice junctions follow a 205-base ladder. Proc Natl Acad Sci U S A. 1991;88:2380–3. doi: 10.1073/pnas.88.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–6. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8.2 ExceptionalEvaluated by Steven Henikoff 07 Jan 2009, Lin Chen 27 Feb 2009, Jesper Svejstrup 02 Apr 2009
- 7.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–43. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 9.0 ExceptionalEvaluated by Paul Liu 29 Apr 2008
- 10.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, Sidow A, Fire A, Johnson SM. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–63. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]; F1000 Factor 7.0 Must ReadEvaluated by Steven Henikoff 22 May 2007, Xing Wang Deng 05 Jun 2007, Michael Meisterernst 19 Jun 2007, Deyou Zheng 29 Jun 2007, Magdalena Zernicka-Goetz 15 Jan 2008
- 12.Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–81. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–5. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 14.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, Schmidt D, O’Keeffe S, Haas S, Vingron M, Lehrach H, Yaspo ML. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Charles Auffray 02 Feb 2009
- 15.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.4 Must ReadEvaluated by Ken Irvine 20 Nov 2008, Donald Rio 01 Dec 2008
- 16.Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, Shai O, Lee L, Rozenhek A, Mohammad N, Willaime-Morawek S, Babak T, Zhang W, Hughes TR, van der Kooy D, Frey BJ, Blencowe BJ. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, Wong WK, Mockler TC. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.2 RecommendedEvaluated by Julin Maloof 17 Feb 2010, Daniel Chamovitz 06 Jul 2010
- 18.Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, Brenner SE. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gan Q, Chepelev I, Wei G, Tarayrah L, Cui K, Zhao K, Chen X. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010;20:763–83. doi: 10.1038/cr.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juneau K, Palm C, Miranda M, Davis RW. High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing. Proc Natl Acad Sci U S A. 2007;104:1522–7. doi: 10.1073/pnas.0610354104. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by Miriam Zolan 09 Jan 2009
- 21.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–43. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 22.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–9. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Anne Baranger 20 Aug 2008
- 23.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11:345–55. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 24.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010;277:856–66. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–13. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001;98:11193–8. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz SH, Silva J, Burstein D, Pupko T, Eyras E, Ast G. Large-scale comparative analysis of splicing signals and their corresponding splicing factors in eukaryotes. Genome Res. 2008;18:88–103. doi: 10.1101/gr.6818908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwartz S, Gal-Mark N, Kfir N, Oren R, Kim E, Ast G. Alu exonization events reveal features required for precise recognition of exons by the splicing machinery. PLoS Comput Biol. 2009;5:e1000300. doi: 10.1371/journal.pcbi.1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–45. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]; F1000 Factor 8.0 ExceptionalEvaluated by Donald Rio 23 Dec 2004, Reinhard Luehrmann 05 Jan 2005
- 30.Schwartz S, Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–36. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tilgner H, Guigo R. From chromatin to splicing: RNA-processing as a total artwork. Epigenetics. 2010;5:180–4. doi: 10.4161/epi.5.3.11319. [DOI] [PubMed] [Google Scholar]
- 32.Kornblihtt AR, Schor IE, Allo M, Blencowe BJ. When chromatin meets splicing. Nat Struct Mol Biol. 2009;16:902–3. doi: 10.1038/nsmb0909-902. [DOI] [PubMed] [Google Scholar]
- 33.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci U S A. 2009;106:4325–30. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alló M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, Klinck R, Chabot B, Kornblihtt AR. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–24. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Andrea Barta 16 Jul 2009
- 35.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.9 Must ReadEvaluated by Andrea Mattevi 17 Feb 2010, Leonie Ringrose 02 Mar 2010, Luc Desgroseillers 24 Mar 2010, Gabriele Varani 06 Apr 2010, Herbert Steinbeisser 28 May 2010
- 36.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–9. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]; F1000 Factor 6.8 Must ReadEvaluated by Bino John 10 May 2010, Igor Stagljar 14 May 2010, Dean Sheppard 24 May 2010, David Bates 03 Jun 2010, Juan Valcárcel 15 Jun 2010
- 37.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–41. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 6.0 Must ReadEvaluated by Reinhard Luehrmann 07 Sep 2009
- 38.Nahkuri S, Taft RJ, Mattick JS. Nucleosomes are preferentially positioned at exons in somatic and sperm cells. Cell Cycle. 2009;8:3420–4. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- 39.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–5. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 40.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–54. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 42.Chen W, Luo L, Zhang L. The organization of nucleosomes around splice sites. Nucleic Acids Res. 2010;38:2788–98. doi: 10.1093/nar/gkq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.0 RecommendedEvaluated by I King Jordan 07 Jan 2010
- 44.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–8. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 3.2 RecommendedEvaluated by Jim Maher 17 Aug 2009, Gabriele Varani 27 Oct 2009
- 45.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–33. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Factor 8.2 ExceptionalEvaluated by Pamela Geyer 04 Nov 2009, Christine Clayton 15 Dec 2009, Jurg Bahler 14 Jan 2010, Miles Wilkinson 11 Jun 2010
- 46.Bert AG, Johnson BV, Baxter EW, Cockerill PN. A modular enhancer is differentially regulated by GATA and NFAT elements that direct different tissue-specific patterns of nucleosome positioning and inducible chromatin remodeling. Mol Cell Biol. 2007;27:2870–85. doi: 10.1128/MCB.02323-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell. 2009;36:541–6. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]