The novel polyI:C-inducible membrane protein INAM triggers dendritic cell–mediated natural killer cell activation.
Abstract
In myeloid dendritic cells (mDCs), TLR3 is expressed in the endosomal membrane and interacts with the adaptor toll/interleukin 1 receptor homology domain–containing adaptor molecule 1 (TICAM-1; TRIF). TICAM-1 signals culminate in interferon (IFN) regulatory factor (IRF) 3 activation. Co-culture of mDC pretreated with the TLR3 ligand polyI:C and natural killer (NK) cells resulted in NK cell activation. This activation was triggered by cell-to-cell contact but not cytokines. Using expression profiling and gain/loss-of-function analyses of mDC genes, we tried to identify a TICAM-1–inducing membrane protein that participates in mDC-mediated NK activation. Of the nine candidates screened, one contained a tetraspanin-like sequence and satisfied the screening criteria. The protein, referred to as IRF-3–dependent NK-activating molecule (INAM), functioned in both the mDC and NK cell to facilitate NK activation. In the mDC, TICAM-1, IFN promoter stimulator 1, and IRF-3, but not IRF-7, were required for mDC-mediated NK activation. INAM was minimally expressed on NK cells, was up-regulated in response to polyI:C, and contributed to mDC–NK reciprocal activation via its cytoplasmic tail, which was crucial for the activation signal in NK cells. Adoptive transfer of INAM-expressing mDCs into mice implanted with NK-sensitive tumors caused NK-mediated tumor regression. We identify a new pathway for mDC–NK contact-mediated NK activation that is governed by a TLR signal-derived membrane molecule.
Natural killer (NK) cells contribute to innate immune responses by killing virus-infected or malignantly transformed cells and by producing cytokines such as IFN-γ and TNF. NK cell activation is determined by a balance of signals from inhibitory and activating receptors. Because ligands of inhibitory receptors include MHC class I and class I–-like molecules, the absence of self-MHC expression leads to NK activation (Cerwenka and Lanier, 2001). Approximately 20 receptors contribute to NK activation (Cerwenka and Lanier, 2001; Vivier et al., 2008). When ligands for activating receptors are sufficiently abundant, activating signals overcome inhibitory signals.
There are two currently accepted models for in vivo NK activation. One is that NK cells usually circulate in a naive state and are activated through interaction directly with ligands for pattern recognition receptors (PRRs) expressed by NK cells or interaction with cells that express PRR ligands (Hornung et al., 2002; Sivori et al., 2004). When pathogens enter the host, innate immune sensors, such as Toll-like receptors (TLRs), RIG-I-like receptors, NOD-like receptors, and lectin family proteins, which are PRRs, recognize a variety of microbial patterns (pathogen-associated molecular patterns [PAMPs]; Medzhitov and Janeway, 1997). Mouse NK cells express almost all TLRs (TLR1–3, 4, and 6–9), and some of these are directly activated by pathogens with the help of IL-12, IL-18, IFN-γ, and other cytokines (Newman and Riley, 2007). The other is that naive NK cells tend to be recruited to the draining LNs, where they are primed to be effectors with the help of mature myeloid DCs (mDC) and released into peripheral tissues (Fernandez et al., 1999). In this case, mDCs provide direct activating signals to NK cells through cell–cell contact (Gerosa et al., 2002; Akazawa et al., 2007a; Lucas et al., 2007). mDCs also produce proinflammatory cytokines and IFN-α after recognizing PAMPs (Newman and Riley, 2007). In this mDC-mediated NK activation, however, the molecules and mechanisms in mDC that are dedicated to NK activation in vivo remain to be understood.
In this study, we focused on the molecules that are induced in mDC during maturation by exposure to double-stranded (ds) RNA and the molecules involved in priming NK cells for target killing (Akazawa et al., 2007a). dsRNA of viral origin and the synthetic analogue polyI:C induce NK activation in concert with mDC in vivo and in vitro (Seya and Matsumoto, 2009). PolyI:C is recognized by the cytoplasmic proteins RIG-I/MDA5 and the membrane protein TLR3, both of which are expressed in mDC (Matsumoto and Seya, 2008). Although RIG-I and MDA5 in the cytoplasm deliver a signal to the adaptor protein IFN promoter stimulator 1 (IPS-1; also known as MAVS, VISA, and Cardif) on the outer membrane of the mitochondria (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005), TLR3 in the endosomal membrane recruits the adaptor protein toll/IL-1 receptor homology domain–containing adaptor molecule 1 (TICAM-1)/TRIF (Oshiumi et al., 2003a; Yamamoto et al., 2003a). Both adaptor proteins activate TBK1 and/or IκB kinase (IKK) ε, which phosphorylate IFN regulatory factor (IRF) 3 and IRF-7 to induce type I IFN (Sasai et al., 2006). We previously showed that the TLR3–TICAM-1 pathway in mDC participates in inducing anti-tumor NK cytotoxicity by polyI:C (Akazawa et al., 2007a). mDC matured with polyI:C can enhance NK cytotoxicity through mDC-NK cell–cell contact (Akazawa et al., 2007a). Therefore, we hypothesized that an unidentified protein is up-regulated on the cell surface of mDC through activation of the TLR3–TICAM-1 pathway, and this protein enables mDC to interact with and activate NK cells. This is the first study identifying an IRF-3–dependent NK-activating molecule, which we abbreviated INAM. INAM is a TICAM-1–inducible molecule on the cell surface of BM-derived DCs (BMDCs) that activates NK cells via cell–cell contact. Our data imply that mDCs harbor a pathway for driving NK activation that acts in conjunction with dsRNA and TLR3.
RESULTS
TICAM-1/IRF-3 signal in BMDCs augments NK activation
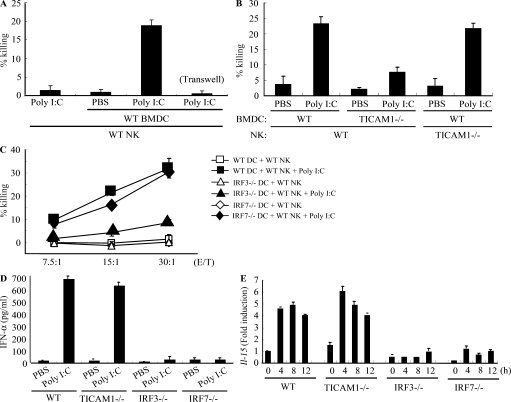
An in vitro system for evaluating NK activation through BMDC-NK contact was established for this study (Fig. 1 A). A mouse melanoma cell subline B16D8, which was established in our laboratory as a low H-2 expressor (Mukai et al., 1999), was used as an NK target. PolyI:C, WT BMDC, and NK cells were all found to be essential for NK-mediated B16D8 cytolysis in the in vitro assay (Fig. 1 A). PolyI:C-mediated NK activation was at baseline levels in a transwell with a 0.4-µm pore, suggesting the importance of direct BMDC-NK contact for this cytolysis induction (Fig. 1 A). When WT BMDCs were replaced with TICAM-1−/− BMDCs in this system, polyI:C-mediated NK activation was partly abolished (Fig. 1 B; and Fig. S1, A and B). TICAM-1 of BMDC was involved in driving NK activation, and ultimately B16D8 cells were damaged by BMDC-derived NK cells (Fig. 1 B). PolyI:C-mediated NK activation occurred even when WT NK cells were replaced with TICAM-1−/− NK cells (Fig. 1 B), which means that NK activation barely depends on the TICAM-1 pathway in NK cells.
Figure 1.
IRF-3 in BMDC controls the capacity to activate NK cells in response to polyI:C. (A and B) WT or TICAM-1−/− NK cells were co-cultured with WT or TICAM-1−/− BMDC in the presence of 10 µg/ml polyI:C for 24 h. NK cytotoxicity against B16D8 was determined by standard 51Cr release assay. E/T = 30. (C) WT NK cells were co-cultured with WT (□, ■), IRF-3−/− (△, ▲), or IRF-7−/− (◇, ◆) BMDC in the presence (■, ▲, ◆) or absence (□, △, ◇) of 10 µg/ml polyI:C for 24 h. NK cytotoxicity against B16D8 was determined by standard 51Cr release assay at the indicated E/T ratio. (D) ELISA of IFN-α in cultures of WT, TICAM-1−/−, IRF-3−/−, and IRF-7−/− BMDC treated with 10 µg/ml polyI:C for 24 h. (E) Quantitative RT-PCR for IL-15 expression in BMDC stimulated with 10 µg/ml polyI:C. All data are means ± SD of duplicate or triplicate samples from one experiment that is representative of three.
PolyI:C-activated splenic NK cells were i.p. injected into B6 mice to kill B16D8 cells ex vivo, which is consistent with previous studies (McCartney et al., 2009; Miyake et al., 2009), and this polyI:C-mediated NK activation was markedly reduced in IPS-1−/− mice established in our laboratory (Fig. S1 C), suggesting that NK cell activation is induced via not only the TICAM-1 pathway but also the IPS-1 pathway, which was largely comparable with previous studies (McCartney et al., 2009; Miyake et al., 2009). IPS-1 in BMDC was more involved in polyI:C-driven NK cytotoxicity than TICAM-1 but almost equally contributed to NK-dependent IFN-γ induction to TICAM-1 in our setting (Fig. S1 B). In addition, the serum level of IL-12p40 in polyI:C-treated mice was largely dependent on TICAM-1 (Fig. S1 D; Kato et al., 2006; Akazawa et al., 2007a). In the supernatant of polyI:C-stimulated BMDC and the serum samples from polyI:C-treated mice, IL-12p70 was not detected by ELISA (unpublished data). These results suggest that polyI:C activates NK cells largely secondary to mDC maturation, which is sustained by the IPS-1 or TICAM-1 pathway of mDC. Even though NK cells express TLR3, they are only minimally activated by polyI:C alone. Signaling by TICAM-1 in BMDC can augment NK cytotoxicity and IFN-γ production via BMDC/NK contact.
The TICAM-1 pathway activates the transcription factor IRF-3. More precisely, exogenous addition of polyI:C can activate endosomal TLR3 and cytoplasmic RIG-I/MDA5. RIG-I/MDA5 assembles the adaptor IPS-1, which in turn recruits the NAP1–IKK-ε–TBK1 kinase complex and activates both IRF-3 and IRF-7 (Fitzgerald et al., 2003; Yoneyama et al., 2004). For this reason, we examined the role of IRF-3 and IRF-7 in BMDC for activation of NK cells by polyI:C. Activation of IRF-3, but not IRF-7, was required for BMDC to induce NK cytotoxicity (Fig. 1 C). IL-2 (Zanoni et al., 2005), IFN-α (Gerosa et al., 2002), and trans-presenting IL-15 (Lucas et al., 2007) induced by BMDC are reported to be key cytokines for BMDC-mediated NK activation in response to polyI:C. However, even with normal levels of IFN-α production and IL-15 expression (Fig. 1, D and E), TICAM-1−/− BMDCs failed to induce full NK cytotoxicity (Fig. 1 B). In contrast, IRF-7−/− BMDCs, which have impaired IFN-α and IL-15 expression, fully activated NK cells (Fig. 1, C–E). Hence, in BDMCs, the TICAM-1–IRF-3 pathway, rather than other cytokines, appears to induce cell surface molecules that mediate BMDC/NK contact and evoke NK cytotoxicity.
Identification of INAM
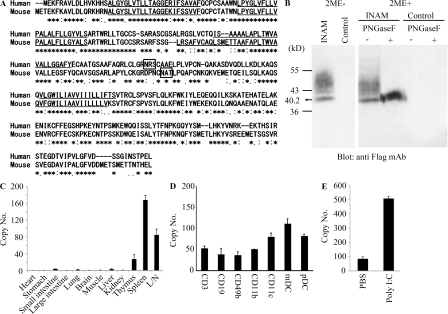
To identify the NK-activating cell surface molecule on BMDC, we performed microarray analysis on polyI:C-stimulated BMDC prepared from TICAM-1−/− and WT mice. The results yielded nine TICAM-1–inducible molecules with transmembrane motifs (Table S1). Six were induced in an IRF-3–dependent manner, whereas three were still induced in IRF-3−/− BMDC. The NK-activating ability of the products of these genes was investigated by introduction of lentivirus expression vector into IRF-3−/− BMDC. BMDCs with the transduced genes were co-cultured with WT NK cells and polyI:C, and the NK-activating ability was evaluated by determining IFN-γ in the 24-h co-culture. NK cells, but not the gene-transduced BMDCs, produced IFN-γ in the presence of polyI:C. Finally, we identified a tetraspanin-like molecule that satisfied our evaluation criteria (IFN-γ and cytotoxicity) on the mDC-NK activation and named this molecule INAM. INAM clearly differed from other tetraspanins like CD9, CD63, CD81, CD82, and CD151 in the predicted structure. Mouse INAM is a 40–55-kD protein with one N-glycosylation site and possesses four transmembrane motifs (Fig. 2, A and B). Western blotting analysis of INAM-transfected cells under nonreducing conditions showed no evidence of multimers (Fig. 2 B). The N-terminal and C-terminal regions of INAM are in the cytoplasm because anti-Flag antibody did not detect C-terminal Flag-tagged INAM until cells were permeabilized (unpublished data).
Figure 2.
Sequence alignment of INAM and expression of INAM. (A) Sequence alignment of human and mouse INAM. Asterisks, identical residues; double dots, conserved substitutions; single dots, semiconserved substitutions; box, N-glycosylation site; underline, transmembrane motif. (B) Immunoblot analysis of lysates of 293FT cells transfected with plasmid encoding Flag-tagged INAM. PNGase, N-glycosidase. 2ME, 2-mercaptoethanol. (C and D) Quantitative RT-PCR for INAM expression in mouse tissue (C) and spleen cells (D). CD3+, CD19+, DX5+, CD11b+, CD11c+, mDC (CD11c+PDCA1+), and plasmacytoid DC (pDC; CD11c+PDCA1+) cells were isolated from splenocytes by cell sorting. Data are expressed as copy number per 104 copies of HPRT. Data shown are means ± SD of triplicate samples from one experiment that is representative of three. (E) Augmented INAM expression in LN cells after polyI:C stimulation. WT mice were i.p. injected with 100 µg polyI:C or control buffer. After 24 h, inguinal, axillary, and mesenteric LN were harvested and RNA was extracted from the LN cells. The levels of the INAM mRNA were measured by real-time PCR. The results were confirmed in two additional experiments. Data represent mean ± SD.
Alignment of the predicted amino acid sequence of mouse INAM with that of the human orthologue revealed that the two INAMs shared a 71.7% amino acid identity. INAM is also called FAM26F (Table S1) and is in the FAM26 gene family (Bertram et al., 2008; Dreses-Werringloer et al., 2008). Sequence database searches identified six mouse INAM paralogs. Although FAM26A/CALHM3, FAM26B/CALHM2, and FAM26C/CALHM1 are located on chromosome 19, FAM26D, FAM26E, and FAM26F/INAM are on chromosome 10. Only INAM was inducible with TLR agonists (unpublished data). All FAM26 family proteins have three or four transmembrane motifs predicted by the TMHMM Server (version 2.0). Human CALHM1 has a conserved region (Q/R/N site) with ion channel properties at the C-terminal end of the second transmembrane motif that controls cytoplasmic Ca2+ levels (Dreses-Werringloer et al., 2008). However, the Q/R/N site was not found in INAM. CALHM1, 2, and 3 are highly expressed in brain. Quantitative RT-PCR revealed that INAM expression was high in spleen and LNs but low in thymus, liver, lung, and small intestine (Fig. 2 C), although expression of the other two FAM26 family members from chromosome 10 was highest in brain (not depicted). All splenocytes examined (CD3+, CD19+, DX5+, CD11b+, CD11c+, mDCs [CD11c+PDCA1−], and plasmacytoid DCs [CD11c−PDCA1+]) expressed INAM to some levels (Fig. 2 D). The INAM expression was inducible by polyI:C in LN cells (Fig. 2 E); the induction levels were more prominent in myeloid cells than in lymphocytes in the LNs (Fig. S2 A). NKp46+ and DX5+ NK cells also expressed INAM with low levels and the levels were mildly increased by polyI:C stimulation (Fig. S2 A and not depicted). Notably, only CD45+ cells expressed INAM, which excludes the participation of contaminating stromal cells in the INAM up-regulation (Fig. S2 B).
BMDC INAM activates NK cells
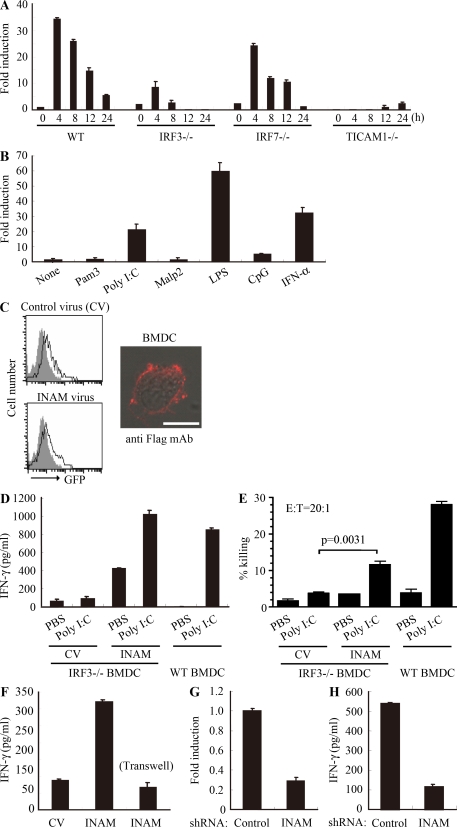
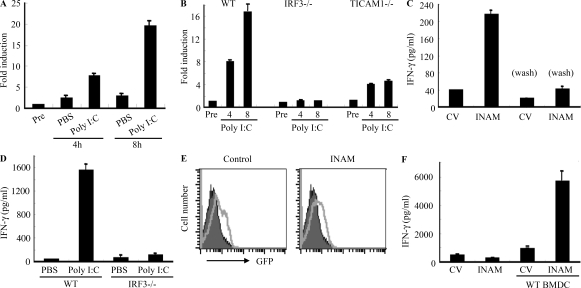
WT and IRF-7−/− BMDCs induced high NK cytotoxicity in response to polyI:C, whereas TICAM-1−/−, IPS-1−/−, and IRF-3−/− BMDC showed less NK activation (Fig. 1, B and C; and Fig. S1). INAM expression profile by polyI:C stimulation was then examined using WT, IRF-3−/−, IRF-7−/−, and TICAM-1−/− BMDCs. Stimulation with polyI:C induced INAM at normal levels in IRF-7−/− BMDC but at decreased levels in IRF-3−/− and TICAM-1−/− BMDC (Fig. 3 A). The expression profiles of INAM in polyI:C-stimulated BMDC were in parallel with those inducing NK activation. BMDCs express a variety of TLRs (Iwasaki and Medzhitov, 2004), but other TLR ligands, Pam3CSK4 for TLR1/2, Malp2 for TLR2/6, and CpG for TLR9, barely induced INAM on BMDC. High induction of INAM was observed in BMDC stimulated with LPS as well as polyI:C (Fig. 3 B), both of which can activate TICAM-1 to induce IRF-3 and IFN-α activation (Fitzgerald et al., 2003; Oshiumi et al., 2003a,b; Yamamoto et al., 2003a,b). Because INAM is an IFN-inducible gene (Fig. 3 B), INAM induction may be amplified by type I IFNs.
Figure 3.
INAM in BMDC participates in DC-mediated NK activation. (A) Quantitative RT-PCR for INAM expression in WT, TICAM-1−/−, IRF-3−/−, and IRF-7−/− BMDC stimulated with 10 µg/ml polyI:C. (B) Quantitative RT-PCR for INAM expression in WT BMDC stimulated by 100 ng/ml LPS, 10 µg/ml polyI:C, 1 µg/ml Pam3, 100 nM Malp-2, 10 µg/ml CpG, and 2,000 IU/ml IFN-α for 4 h. (C) BMDCs were transduced with Flag-tagged INAM-expressing lentivirus or control lentivirus. GFP expression in the BMDC was determined by flow cytometry, and subcellular localization of INAM was examined by immunofluorescence assay using anti-Flag mAb. Shaded peak, noninfected control; Blank peak, infected BMDC. Bar, 10 µm. (D) ELISA of IFN-γ induced by WT NK cells co-cultured with WT BMDC or IRF-3−/− BMDC transfected with control lentivirus (CV) or INAM-expressing lentivirus (INAM) with/without 10 µg/ml polyI:C. (E) Cytotoxicity against B16D8 by NK cells co-cultured with BMDC transfected with control or INAM-expressing lentivirus with/without 10 µg/ml polyI:C for 24 h. (F) ELISA of IFN-γ induced by WT NK cells co-cultured with IRF-3−/− BMDC transfected with control lentivirus (CV) or INAM-expressing lentivirus (INAM) with 10 µg/ml polyI:C. In some experiments, a transwell was inserted between the INAM-transduced BMDC and NK cells to separate the cells. (G) Quantitative RT-PCR for expression of INAM in BMDC transduced with INAM-shRNA (INAM) or scrambled shRNA (control) and cultured for 48 h. (H) IFN-γ production by WT NK cells determined using ELISA after coculturing with control or the shRNA transfected-BMDC (INAM) and 10 µg/ml polyI:C for 24 h. All data shown are means ± SD of triplicate samples from one experiment that is representative of three.
We next examined whether INAM was localized to the cell surface membrane in BMDC. Immunofluorescence analysis showed Flag-tagged INAM on the cell surface of BMDC. Plasma membrane expression of INAM was also confirmed by cell surface biotinylation (Fig. S3). Although the lentivirus inefficiently infected BMDC, GFP expression levels were similar in cells with control virus and those with INAM-expressing virus (Fig. 3 C). Transduction efficiency and expression from the lentivirus vector were adjusted using GFP expression (not depicted), and surface INAM expression was further confirmed with BMDC, NK cells, and INAM-expressing BaF3 (INAM/BaF3) cells, in some cases using polyclonal antibody (Ab) against INAM (Fig. S4).
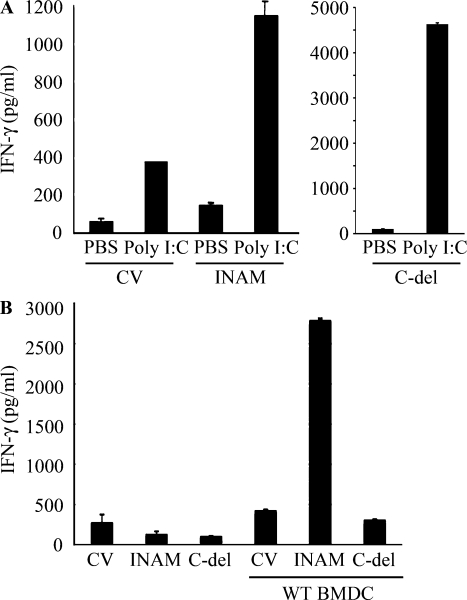
We then examined whether overexpressing INAM resulted in signaling that directed BMDC maturation and production of cytokines, including IFN-α and IL-12p40, which are reported to enhance NK activity (Gerosa et al., 2002; Sivori et al., 2004; Lucas et al., 2007). The status of INAM-transduced BMDC was assessed by CD86 expression and cytokine production, and no significant differences in these maturation markers were seen in BMDC overexpressing INAM (Fig. S5). In the same setting, no IL-12p70 was detected by ELISA (unpublished data). In addition, polyI:C-mediated NK activation occurred in BMDC expressing an INAM mutant lacking the cytoplasmic C-terminal region (193–327 aa; Fig. 4, A and B), excluding the participation of the cytoplasmic region in BMDC maturation signaling.
Figure 4.
Role of the cytoplasmic tail of INAM. (A) The C-terminal region of INAM was not required for BMDC-mediated NK activation. ELISA of IFN-γ by WT NK cells co-cultured with IRF-3−/− BMDCs transfected with control lentivirus (CV) or a lentivirus expressing intact INAM or a mutant INAM lacking the C-terminus (C-del INAM) with/without 10 µg/ml polyI:C. Data shown are means ± SD of triplicate samples from one experiment representative of three. (B) The cytoplasmic tail of INAM is indispensable for NK IFN-γ induction. INAM or C-del INAM (A) was expressed on IRF-3−/− NK cells. The INAM (or C-del INAM)-expressing IRF-3−/− NK cells were incubated with or without WT BMDC for 24 h. IFN-γ levels in the supernatants were determined by ELISA. One representative result out of several similar experiments is shown. Data represent mean ± SD.
To investigate whether INAM could reconstitute NK-activating ability in IRF-3−/− BMDC, we transduced INAM into IRF-3−/− BMDC and incubated BMDC with NK cells. Overexpression of INAM in IRF-3−/− BMDC induced NK IFN-γ production and NK cytotoxicity against B16D8, and this NK activation was further enhanced by the addition of polyI:C (Fig. 3, D and E). Thus, polyI:C may also work for NK activation. Direct cell–cell contact with NK cells was required for INAM in IRF-3−/− BMDC to function on enhancing NK activity (Fig. 3 F).
We further confirmed this issue using WT BMDC by shRNA gene silencing. We silenced the INAM gene in BMDC using the lentiviral vector pLenti-dest-IRES-hrGFP and monitored expression by GFP. Because transfection efficiency was relatively high in this case compared with that shown in Fig. 3 C, the expression level of INAM had decreased by ∼75% in WT BMDC compared with the nonsilenced control (Fig. 3 G and Fig. S6 A). Although the level of the endogenous INAM protein was not very high, we confirmed that INAM protein was also decreased by shRNA with immunoblotting using anti-INAM pAb (Fig. S7 A). PolyI:C response of BMDC-inducible cytokines tested was not altered by INAM silencing in BMDC (Fig. S6 B). Yet this INAM RNA interference caused a significant decrease in NK cell IFN-γ production after co-culture of the INAM knockdown BMDCs and WT NK cells with polyI:C (Fig. 3 H). Collectively, these results indicate that INAM is downstream of IRF-3 in BMDC and is involved in the activation of NK cells by BMDC.
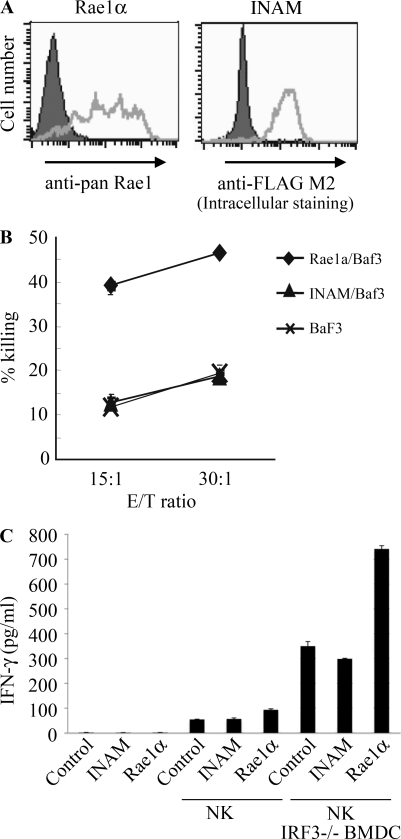
Using an INAM-expressing stable BaF3 cell line (INAM/BaF3), we tested the possibility that INAM is an activating ligand for NK cells. As a positive control, we produced a stable BaF3 cell line expressing Rae-1α (Fig. 5 A) which is a ligand for the NK-activating receptor NKG2D (Cerwenka et al., 2000). Although Rae-1α/BaF3 cells were easily damaged by IL-2–activated NK cells, INAM/BaF3 cells were not (Fig. 5 B). In this context, addition of IRF-3−/− BMDC to this culture with BaF3 and NK cells led to slight augmentation of IFN-γ induction irrespective of the presence of INAM on BaF3 cells (Fig. 5 C), and β2-microglobulin−/− BMDC barely affected the IFN-γ level (not depicted). These results suggest that an INAM-containing molecular matrix, rather than INAM alone, acts toward NK cells. Alternatively, INAM may selectively function with specific mDC molecules to activate NK cells.
Figure 5.
INAM is not an NK-activating ligand. (A) Flow cytometry for Rae-1 and Flag-tagged INAM in stable BaF3 lines. Shaded peak, untransfected control BaF3 staining with anti–pan-Rae-1 Ab or anti-Flag M2 antibody; open peak, stable Rae-1α/BaF3 or stable Flag-tagged INAM/BaF3 staining with anti–pan-Rae-1 antibody or anti-Flag M2 antibody. (B) Cytotoxicity against control BaF3, Rae-1α/BaF3, and INAM/BaF3 by NK cells treated with 1,000 IU/ml IL-2 for 3 d. Data shown are means ± SD of triplicate samples from one experiment representative of three. (C) NK activation is augmented by coexistent BMDC irrespective of INAM expression. NK cells were cultured with 1,000 IU/ml IL-2 for 3 d. 2 × 105 NK cells, 105 BaF3 cells, and 105 IRF-3−/− BMDCs were co-cultured in 200 µl/well and IFN-γ in the supernatants were measured by ELISA. Data show one of two similar experimental results. Data represent mean ± SD.
INAM on NK cells is required for efficient NK activation
mDCs were previously shown to be required for efficient NK activation in vivo and in vitro (Akazawa et al., 2007a). We found that INAM was minimally present in BMDCs and NK cells and that polyI:C acts on both (Figs. S2 A; and Fig 3, D and E). Tetraspanin-like molecules tend to work as scaffolds for heteromolecular complexes that contain molecules functioning in a cis- or trans-adhesion manner to exert intercellular or extracellular functions. Thus, the function of INAM may not be confined to mDC, so we studied the function of INAM on NK cells. In NK cells, INAM was also inducible by polyI:C (Fig. 6 A and Fig. S2 A), and the induction of INAM was abrogated completely in IRF-3−/− NK cells and moderately in TICAM1−/− NK cells (Fig. 6 B). This suggests that polyI:C also acts on NK cells and induces INAM through IPS-1/IRF-3 activation when NK cells are co-cultured with BMDC and polyI:C.
Figure 6.
INAM on NK cells contributes to efficient NK activation mediated by mDC. (A and B) Quantitative RT-PCR for INAM expression in WT, TICAM1−/−, or IRF-3−/− NK cells stimulated with 50 µg/ml polyI:C. Data shown are means of duplicate or triplicate samples from one experiment that is representative of three. (C) IRF-3−/− BMDCs were transfected with control lentivirus (CV) or INAM-expressing lentivirus (INAM) before treatment with 10 µg/ml polyI:C for 4 h. BMDCs in some wells were washed to remove polyI:C before WT NK cells were added (Wash). IFN-γ production by NK cells was determined by ELISA after 24 h of culture. Data show one of two similar experimental results. (D) ELISA of IFN-γ in co-culture of WT or IRF-3−/− NK cells and WT BMDC with/without 10 µg/ml polyI:C. (E and F) NK cells were transfected with control lentivirus or INAM-expressing lentivirus and cultured with 500 IU/ml IL-2 for 3 d. After determining transfection efficiency by GFP intensity using flow cytometry, cells were cultured with/without BMDC for 24 h and IFN-γ production in the supernatant determined by ELISA. Shaded peak, noninfected control; open peak, infected BMDC. All data are means ± SD of triplicate samples from one experiment that is representative of three.
To investigate whether INAM induced in NK cells is associated with BMDC-mediated NK activation, we performed the following experiments (Fig. 6 C). INAM-transduced IRF-3−/− BMDCs were incubated with polyI:C for 4 h, and then the aliquot was mixed with WT NK cells in the presence of polyI:C (Fig. 6 C, left two lanes). A moderate increase of IFN-γ was observed as in Fig. 3 D. In the remainder, we washed polyI:C out and cultured the cells with WT NK cells (Fig. 6 C, right two lanes). Under these conditions, in which polyI:C acted not on NK cells but only on BMDC, little NK activation was observed (Fig. 6 C). Furthermore, IRF-3−/− NK cells produced little IFN-γ when co-cultured with WT BMDC and polyI:C (Fig. 6 D). INAM-overexpressing IRF-3−/− BMDC required IRF-3 in NK cells for efficient BMDC-mediated production of IFN-γ from NK cells (Fig. 6 D). We next transduced INAM into IRF-3−/− NK cells using a lentivirus (INAM/pLenti-IRES-hrGFP) to reconstitute NK IFN-γ–producing activity. After many trials with various setting conditions, we found that ∼15% of the DX5+ NK cell population was both GFP-positive and stained with anti-FLAG mAb when treated with high doses of INAM-expressing lentivirus vector (Fig. S7 B). When IRF-3−/− NK cells were infected with smaller amounts of INAM-expressing lentiviral vector and cultured for 3 d with high concentrations of IL-2 (500 IU/ml), slight but significant GFP expression was confirmed by FACS (Fig. 6 E). Then, the INAM-transduced IRF-3−/− NK cells were co-cultured with WT BMDC. The IRF-3−/− NK cells with INAM expression secreted IFN-γ at significantly higher levels than controls in the presence of WT BMDC (Fig. 6 F). These data indicate that INAM is induced by polyI:C through IRF-3 activation, not only in BMDCs but also in NK cells, and that INAM on NK cells synergistically works with INAM on BMDC for efficient NK cell activation. Both INAMs on BMDC and NK cells are essential for BMDC-mediated NK activation.
We next checked the function of the C-terminal stretch of INAM in NK activation. Although intact INAM works in NK cells to produce IFN-γ in response to BMDC (Fig. 6 F), introduction of C-del INAM into IRF-3−/− NK cells did not result in high induction of IFN-γ in response to BMDC (Fig. 4 C). Thus, INAM participates in NK activation through its cytoplasmic regions, which has no significant role in BMDC for NK activation.
Anti-tumor NK activation via INAM-expressing BMDCs in vivo
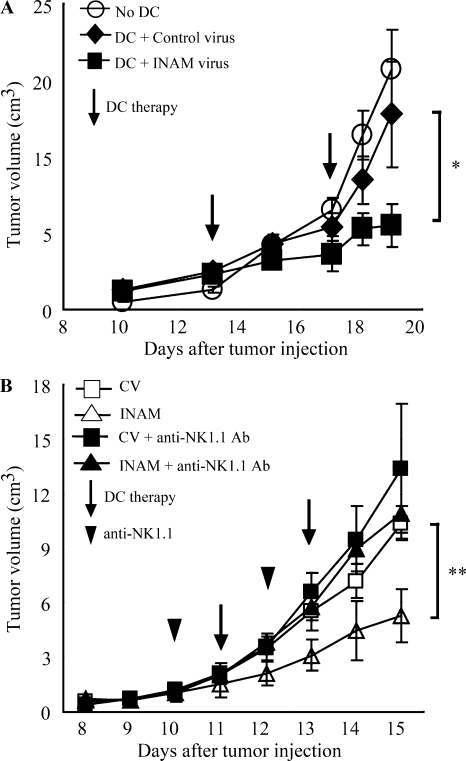
mDC-mediated NK activation induces anti-tumor NK cells, which cause regression of NK-sensitive tumors (Kalinski et al., 2005; Akazawa et al., 2007a). We tested the in vivo function of INAM-expressing BMDC using B16D8 tumor-bearing mice. BMDCs were used 24 h after transfection with either INAM/pLenti-IRES-hrGFP or control pLenti-IRES-hrGFP and injected twice a week s.c. around a preexisting tumor in tumor-implanted mice, beginning 11–13 d after tumor challenge. INAM-expressing BMDC significantly retarded tumor growth (Fig. 7 A). Tumor retardation was abrogated by depletion of NK1.1-positive cells (Fig. 7 B). Thus, INAM expression on BMDC contributed to anti-tumor NK activation in vivo.
Figure 7.
INAM on BMDC retarded B16D8 tumor growth in an NK-dependent manner. (A) Tumor volume after DC therapy using BMDC expressing INAM. B16D8 cells were s.c. injected into C57BL/6 mice and, 11–13 d later, medium only (○) or BMDC (106/mouse) transfected with control lentivirus (◆) or those with INAM-expressing lentivirus (■) were administered s.c. near the tumor at the time indicated by the open arrow. *, P = 0.043. Data represent mean ± SD. (B) Abrogation of INAM-dependent tumor regression by administration of NK1.1 Ab. For depletion of NK cells, antiNK1.1 mAb was injected i.p. 1 d before treatment of BMDC (arrowheads). Tumor volume in every mouse group was sequentially monitored. Data represent mean ± SD (n = 3) and are representative of two experiments. Statistical analyses were made with the Student’s t test. **, P = 0.017.
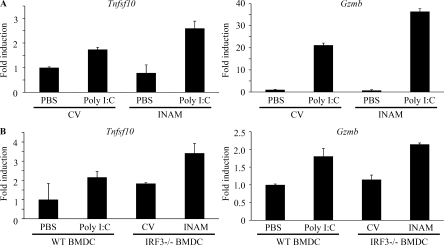
When the control or INAM-expressing IRF-3−/− BMDCs were co-cultured with WT NK cells in vitro, there was no induction of the mRNA of TRAIL and granzyme B in NK cells (Fig. 8 A). TRAIL and granzyme B were induced in NK cells by the addition of polyI:C to the mixture, and INAM expression in BMDC up-regulated mRNA levels of TRAIL and granzyme B (Fig. 8 A). In vivo administration studies were performed with polyI:C-treated WT BMDC or INAM-expressing IRF-3−/− BMDC to test their ability to up-regulate the mRNA levels of TRAIL and granzyme B in NK cells in draining LN (Fig. 8 B). INAM-expressing IRF-3−/− BMDC showed comparable abilities to up-regulate the killing effectors with polyI:C-treated BMDC (Fig. 8 B). Collectively, INAM has therapeutic potential for NK-sensitive tumors by activating NK cells.
Figure 8.
INAM-mediated induction of TRAIL and granzyme B in BMDC. (A) In vitro induction of TRAIL (Tnfsf10) and granzyme B (Gzmb) mRNA by INAM-expressing BMDC. BMDCs (IRF-3−/−) were infected with INAM-expressing virus or CV as in Fig. S4. After 24 h, the BMDCs (IRF-3−/−) were incubated with WT NK cells at DC/NK = 1:2. 8 h later, DX5+ cells were collected by FACS sorting and their RNA was extracted to determine the mRNA levels of the indicated genes. A representative result of three similar experiments are shown. (B) In vivo induction of TRAIL and granzyme B mRNA by INAM-expressing BMDC. WT BMDCs were stimulated with 10 µg/ml polyI:C or medium only. IRF-3−/− BMDCs were infected with CV or INAM-expressing vector. These BMDCs were allowed to stand for 24 h and then 5 × 105 cells were injected into footpads of WT mice. After 48 h, DX5+ cells were collected from the inguinal LN by FACS sorting. RNA of the cells was extracted and the levels of the indicated mRNA were determined by real time PCR. Data show one of two experiments with similar results. Data in A and B represent mean ± SD.
DISCUSSION
Previous studies demonstrated that mDC–NK interaction leads to direct NK activation and damages NK target cells in vitro (Gerosa et al., 2002; Sivori et al., 2004; Akazawa et al., 2007a; Lucas et al., 2007). In addition, mDCs initiate NK cell–mediated innate anti-tumor immune responses in vivo (Kalinski et al., 2005; Akazawa et al., 2007a,b). Systemic administration of polyI:C unequivocally results in activation of peripheral NK cells (Lee et al., 1990; Sivori et al., 2004; Akazawa et al., 2007a). Although the molecular mechanism by which mDCs prime NK cells was still unclear, the TICAM-1 pathway and IPS-1 pathway have been reported to participate in polyI:C-mediated mDC maturation that drives NK activation (Akazawa et al., 2007a; McCartney et al., 2009; Miyake et al., 2009). We have shown in an earlier study that mDCs disrupted in the TLR3–TICAM-1 pathway abrogate NK cell activation (Akazawa et al., 2007a,b). In TICAM-1−/− mice, NK-sensitive implant tumors grew as well as those in WT mice depleted of NK cells (Akazawa et al., 2007a). mDCs gain high anti-tumor potential against B16D8 implant tumors through lentiviral transfer of TICAM-1, which is attributable to NK activation (Akazawa et al., 2007a). We further showed that TICAM-1 is a critical molecule for mDC to induce NK cell IFN-γ, as well as IPS-1, and participates in driving NK cytotoxicity to a lesser extent than IPS-1. In this paper, we clarified a molecular mechanism by which mDCs immediately promote NK cell functions in vitro and in vivo.
Our findings showed that IRF-3 is the transcription factor that is downstream of TICAM-1 responsible for maturing mDC to an NK-activating phenotype. We discovered that INAM, a membrane-associated protein, is up-regulated on the surface of mDC by polyI:C stimulation and activates NK cells via cell–cell contact. Furthermore, we found that NK cells also express INAM on their cell surface after polyI:C stimulation. mDC–NK activation by polyI:C can be reproduced with INAM-transduced mDC and NK cells, and adoptive transfer experiments show that INAM-overexpressing mDC may have therapeutic potential against MHC-low melanoma cells in an NK-dependent manner. These functional properties of INAM-expressing mDC fit the model of mDC priming NK activation. Ultimately, INAM appears to be the key molecule in the previously reported mechanism of mDC–NK contact activation.
After the submission of this manuscript, two papers were published that found that the MDA5–IPS-1 pathway in mDC is more important for driving NK activation, particularly in vivo (McCartney et al., 2009; Miyake et al., 2009). Our data also support this point using the IPS-1−/− mice we established (Fig. S1). However, polyI:C, when i.v. administered into mice, may stimulate other systemic cells in addition to CD8+ mDC in vivo (McCartney et al., 2009). The difference among the two (McCartney et al., 2009; Miyake et al., 2009) and this study may be attributed to the setting conditions, which are not always comparable. Moreover, it remains to be settled whether TICAM-1 and IPS-1 take the same INAM complex as a common NK activator in mature mDC and whether TLR3 (or MDA5) KO is equivalent to TICAM-1 (or IPS-1) KO in the mDC–NK activation model. In either case, however, up-regulation of mDC TICAM-1–mediated NK cytotoxicity and IFN-γ induction are feasible with polyI:C under three different conditions (Akazawa et al., 2007a; McCartney et al., 2009; Miyake et al., 2009). Our results infer that INAM participates in at least these mDC–NK interactions.
PolyI:C activates IRF-3 through the two pathways involving the adaptors IPS-1 and TICAM-1 (Yoneyama et al., 2004; Kato et al., 2006; Matsumoto and Seya, 2008). The two pathways share the complex of IRF-3–activating kinase, NAP1, IKK-ε, and TBK1 that is downstream of adaptors (Sasai et al., 2006). Nevertheless, these pathways are capable of inducing several genes unique to each adaptor. Although IFN-α production by in vivo administration of polyI:C is largely dependent on the IPS-1 pathway, IL-12p40 is mainly produced by the TICAM-1 pathway (Kato et al., 2006). Therefore, it is not surprising that INAM induction is predominant in the TICAM-1 pathway in polyI:C-stimulated BMDC (Fig. 3 A). What happens in IRF-7−/− BMDCs in terms of INAM induction and what mechanism sustains BMDC IPS-1–mediated or MyD88-mediated activation of NK cells (Azuma et al., 2010) will be issues to be elucidated in the future.
Although IRF-3–regulated cell surface INAMs are required for efficient interaction between BMDC and NK cells, the mechanism by which forced expression of INAM causes signaling for BMDC maturation is still unknown. Although the NK-activating capacity of BMDCs is usually linked to their maturation, neither cytokines in NK activation, including IFN-α and IL-12p70, nor costimulators, such as CD40 and CD86, were specifically induced in mDC by INAM expression (Fig. S5). INAM has a C-terminal cytoplasmic stretch (Fig. 4 A), and we tested the function of this region by a deletion mutant (C-del INAM). This region in BMDC barely participates in driving NK activation because no decrease of IFN-γ induction by NK cells was observed with IRF-3−/− BMDC supplemented with C-del INAM compared with control INAM. Thus far, no significant signal alteration has been detected in BMDC supplemented with INAM by lentivirus.
In contrast, INAM-transduced IRF-3−/− NK cells produced IFN-γ in concert with BMDCs like WT NK cells (Fig. 6 F). So far we have no evidence suggesting that this kind of INAM overexpression is actually occurring in vivo. However, introduction of C-del INAM into IRF-3−/− NK cells did not result in high induction of IFN-γ in response to BMDC (Fig. 4 C). Together with the data on INAM expression in BMDC, this infers that the INAM cytoplasmic region signals for NK activation in NK cells. The one-way role of the cytoplasmic tail in NK activation will be an issue for further analysis.
In this study, IL-15 was found to be up-regulated by polyI:C in BMDC. The remaining NK activity in the resting population of NK cells co-cultured with TICAM-1−/− BMDC and polyI:C (Fig. 1 B) suggests that IL-15 has some effect in our system, and other studies suggest this as well (Ohteki et al., 2006; Brilot et al., 2007; Lucas et al., 2007; Huntington et al., 2009). However, we did not observe decreased IL-15 expression in the TICAM-1−/− BMDC that could not activate NK cells (Fig. 1 E). Several molecules, such as B7-H6/NKp30 (Brandt et al., 2009), CD48/2B4 (Kubin et al., 1999), and NKG2D ligands/NKG2D (Cerwenka et al., 2000), have been identified as ligand/receptor molecules in mDC–NK reciprocal activation by in vitro co-culture. In in vitro co-culture systems (Fig. S1), the IPS-1 pathway in BMDC has a pivotal role in not only type I IFN but also IL-15 induction. INAM identified in this paper serves a unique function in the in vivo induction of NK activation and may offer a tool to investigate the reported mDC-mediated NK activation.
Rae-1 was reported as a molecule with MHC-like structure (Zou et al., 1996) and later identified as a mouse NKG2D ligand (Cerwenka et al., 2000). Although Rae-1 is a GPI-anchored protein with no cytoplasmic sequences (Nomura et al., 1996), it can act as an NK-activating ligand (Cerwenka et al., 2000, 2001; Masuda et al., 2002). Mouse BaF3 cells become NK-sensitive after forced expression of Rae-1α (Masuda et al., 2002). Actually, mouse macrophages induce Rae-1 expression in response to TLR stimuli (Hamerman et al., 2004). In contrast, INAM-expressing stable BaF3 cell lines (INAM/BaF3) did not reveal a function as an NK cell–activating ligand. NK cell cytotoxicity is directed against Rae-1α/BaF3 cells but not against INAM/BaF3 cells (Fig. 5). Therefore, INAM does not represent a typical NK cell–activating ligand. For NK activation, INAM on BMDC appears to require other molecules that are expressed in BMDC but not in BaF3.
INAM has four transmembrane regions, similar to the cell adhesion tetraspanins, which may support cell–cell contact (Levy and Shoham, 2005). Tetraspanins provide a scaffold that facilitates complex formation with associated proteins. INAM on BMDC and NK cells may use cell–cell interaction to assemble in a synaptic formation to activate NK cells. Because the protein constituents of the tetraspanin complexes are cell specific, we are interested in finding partners for INAM that might participate in efficient BMDC–NK interaction. TLR-inducible cell–cell contact may occur through INAM in an immune cell–specific manner. Gene disruption of this INAM will facilitate clarifying this issue. The identification of INAM defines a novel pathway in mDC–NK reciprocal interaction. This study will lead to further research on the molecules that form complexes on BMDC and NK cells to facilitate BMDC–NK interaction.
MATERIALS AND METHODS
Mice.
All mice were backcrossed with C57BL/6 mice more then seven times before use. TICAM-1−/− (Akazawa et al., 2007a) and IPS-1−/− mice were generated in our laboratory. IRF-3−/− (Sato et al., 2000) and IRF-7−/− mice (Honda et al., 2005) were provided by T. Taniguchi (University of Tokyo, Tokyo, Japan). All mice were maintained under specific pathogen-free conditions in the animal facility of the Hokkaido University Graduate School of Medicine. Animal experiments protocols and guidelines were approved by the Animal Safety Center, Hokkaido University, Japan.
Cells.
The B16D8 cell line was established in our laboratory as a subline of B16 melanoma (Tanaka et al., 1988). This subline was characterized by its low or virtually no metastatic properties when injected s.c. into syngeneic C57BL/6 mice. B16D8 was cultured in RPMI 1640/10% FCS. The mouse B cell line BaF3 was obtained from American Type Culture Collection and cultured in RPMI 1640/10% FCS/2 µM 2ME/5 ng/ml IL-3. Mouse NK cells (DX5+ cell) were positively isolated with MACS Beads (Miltenyi Biotec). Mouse BMDCs were prepared as previously reported (Akazawa et al., 2007a).
For purification of cells from spleen or LN, these tissues were treated with 400 IU MandleU/ml collagenase D (Roche) at 37°C for 25 min in HBSS (Sigma-Aldrich). Then EDTA was added, and the cell suspension was incubated for an additional 5 min at 37°C. After removal of RBC with ACK lysis buffer, splenocytes and LN cells were stained with CD45-FITC, CD3ε-PE, CD19-PE, DX5-PE, CD11b-FITC (eBioscience), and CD11c-FITC (BioLegend) and sorted by a FACSAria II (BD). The purity of sorted cells were >96%.
Construction and expression.
Mouse INAM cDNA (A630077B13Rik) was obtained from RIKEN and placed into expression vector pEFBOS and pLenti-IRES-hrGFP, both of which provide the specialized components needed for expression of a recombinant C-terminal FLAG fusion (Akazawa et al., 2007a). For construction of shRNA-expressing lentivirus vector, The ClaI–XhoI fragment of pLenti6-blockit-dest (Invitrogen) was inserted into pLenti-IRES-hrGFP at the site of ClaI and XhoI. This vector was named pLenti-dest-IRES-hrGFP (pLDIG). INAM sequence 5′-CTTCTCTCCGGTTAGTTATCT-3′ was targeted for INAM knockdown (shINAM/pLDIG) and 5′-AGTCTGACATACTTATACTTA-3′ was used for negative control (shCont/pLDIG). We used a gene-expression kit, Lentiviral system (Invitrogen), as previously described (Akazawa et al., 2007a). Four plasmids (one of the pLenti vectors, pLP1, pLP2, and pLP/VSVG) were transfected into 293 FT packaging cells, and the viral particles for transfection were prepared according to the manufacturer’s protocol. The 100× concentrated virus particles were produced after centrifugation of 8,000 g at 4°C for 16 h. Lentivirus produced by pLenti-IRES-hrGFP and pLDIBG could be tittered by GFP expression using flow cytometry. Because the lentivirus vector pLenti-IRES-hrGFP has the IRES-GFP region, we prepared negative control virus by pLenti-IRES-hrGFP without construct. Infection efficiency for BMDC was high with the control vector compared with the INAM-expressing lentivector (Fig. S6 A).
Real-time PCR.
BMDCs were harvested after 4 h of stimulation by 100 ng/ml LPS, 50 µg/ml polyI:C, 1 µg/ml Pam3CSK4 (Pam3), 100 nM mycoplasmal macrophage-activating lipopeptide-2 (Malp-2), 10 µg/ml CpG, and 2,000 IU/ml IFN-α (Ebihara et al., 2007). Mouse tissues (heart, stomach, small intestine, large intestine, lung, brain, muscle, liver, kidney, thymus, and spleen) were collected from C57BL/6. Splenocytes were stained with CD3-PE, CD19-PE, DX5-PE, CD11b-PE, CD11c-FITC, and PDCA1-PE (eBioscience) and sorted by FACSAria (BD). Purity was >98% in each population. For RNA extraction, we used the RNeasy kit (Invitrogen). After removal of genomic DNA by treatment with DNase, randomly primed cDNA strands were generated with Moloney mouse leukemia virus reverse transcription (Promega). RNA expression was quantified by quantitative RT-PCR with gene-specific primers (IL-15 forward, 5′-TTAACTGAGGCTGGCATTCATG-3′; IL-15 reverse, 5′-ACCTACACTGACACAGCCCAAA-3′; INAM forward, 5′-CAACTGCAATGCCACGCTA-3′; INAM reverse, 5′-TCCAACCGAACACCTGAGACT-3′; β-actin forward, 5′-TTTGCAGCTCCTTCGTTGC-3′; β-actin reverse, 5′-TCGTCATCCATGGCGAACT-3′; HPRT forward, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′; and HPRT reverse, 5′-GAAGGGTAGGCTGGCCTATAGGCT-3′) and values were normalized to the expression of β-actin mRNA or HPRT mRNA.
Other primers for PCR were designed using Primer Express software (Applied Biosystems) for another experiment. The following primers were used for PCR: β-actin forward, 5′-CCTGGCACCCAGCACAAT-3′ and reverse, 5′-GCCGATCCACACGGAGTACT-3′; granzyme B forward, 5′-TCCTGCTACTGCTGACCTTGTC-3′ and reverse, 5′-ATGATCTCCCCTGCCTTTGTC-3′; IFN-α4 forward, 5′-CTGCTGGCTGTGAGGACATACT-3′ and reverse, 5′-AGGCACAGAGGCTGTGTTTCTT-3′; TRAIL (Tnfsf10) forward, 5′-CTTCACCAACGAGATGAAGCAG-3′ and reverse, 5′-TCCGTCTTTGAGAAGCAAGCTA-3′; and IL-12p40 (Il12b), forward, 5′-AATGTCTGCGTGCAAGCTCA-3′ and reverse, 5′-ATGCCCACTTGCTGCATGA-3′.
Anti-INAM pAb.
C-terminal INAM (cINAM; 191–314 aa) was subcloned between the NdeI and Sal1 sites of pColdI vector (Takara Bio Inc.). 6× His-tagged cINAM protein was expressed in BL21 by manufacturer’s methods. The cells were sonicated in 20 mM Tris-HCl, 150 mM NaCL, 1 mM PMSF, and 7 M Urea, pH 7.4, on ice. Expression products of cINAM were purified using the HisTrap HP kit (GE Healthcare). The extracted proteins were refolded by stepwise dialysis against decreasing amounts of urea. Rabbit anti-cINAM polyclonal Ab was produced with the cINAM proteins by standard protocol. IgG was purified by precipitation with 33% ammonium sulfate, dialyzed against PBS.
Surface labeling with biotin.
Biotinylation of cell surface proteins was performed according to the reported method (Tsuji et al., 2001). In brief, ∼108 cells were suspended in 1 ml Hepes-buffered saline (HBS), pH 8.5, and incubated with 10 ml of 10 mg/ml NHS-sulfobiotin (Vector Laboratories) for 1 h at room temperature. Cells were washed in HBS three times and then solubilized with lysis buffer containing 1% NP-40, pH 7.4. The cell lysate was immunoprecipitated with avidin-labeled Abs as described previously (Tsuji et al., 2001).
Immunoblot analysis.
Lysates were harvested 24 h after transfection of Flag-tagged INAM/pEFBOS into 293FT cells and treated with N-glycosidase F (PNGaseF; New England Biolabs, Inc.) by the manufacturer’s method in some experiments. Protein samples were separated on SDS-PAGE and immunoblotted by anti-Flag M2 Ab (Sigma-Aldrich). In some experiments, we used highly purified rabbit anti–mouse INAM polyclonal Ab for immunoblotting. The anti-INAM IgG was further purified with protein A–Sepharose and absorbed with BL21 bacterial lysate (where the INAM immunogen was produced) that contained no INAM peptide.
Confocal microscopy.
BMDCs and NK cells were infected with control or INAM-expressing lentivirus as described previously (Akazawa et al., 2007a). 24 h later, cells were fixed with 4% paraformaldehyde for 30 min and permeabilized with PBS containing 0.5% saponin for 30 min at room temperature. Fixed cells were stained with anti-FLAG mAb and Alexa Fluor 568–conjugated secondary Ab. Stable Ba/F3 transfectants expressing INAM were treated with Cytofix/Cytoperm (BD) according to the manufacturer. Then cells were stained with PE-phalloidin and rabbit anti-INAM pAb followed by Alexa Fluor 488–conjugated secondary Ab. Cells were analyzed on a confocal microscope (LSM 510 META; Carl Zeiss, Inc.) for the detection of INAM.
BMDC–NK interaction.
BMDCs were co-cultured with freshly isolated NK cells (BMDC/NK = ∼1:2–1:5) with or without 10 µg/ml polyI:C for 24 h (Akazawa et al., 2007a). In some experiments, function of BMDCs and NK cells was modified by lentivirus vector before BMDC/NK co-culture. IRF-3−/− BMDCs were transfected by control lentivirus and INAM-expressing lentivirus (INAM/pLenti-IRES-hrGFP) and incubated with 6 µg/ml polybrene for 24 h before co-culture. WT BMDCs were transfected with shRNA-expressing lentivirus (shCont/pLDIG or shINAM/pLDIG) and incubated with 6 µg/ml polybrene for 48 h before co-culture. Freshly isolated NK cells were transfected with control lentivirus and INAM-expressing lentivirus (INAM/pLenti-IRES-hrGFP) and cultured with 6 µg/ml polybrene in the presence of 500 IU/ml IL-2 for 72 h before co-culture. Activation of NK cells was assessed by concentration of IFN-γ (ELISA; GE Healthcare) in the medium and by NK cytotoxicity against B16D8. Cytotoxicity was determined by standard 51Cr release assay as described previously (Akazawa et al., 2007a).
Ex vivo NK activation.
Mice were i.p. injected with 250 µg polyI:C. After 24 h, spleen cells were harvested and then NK cells (DX5+ cells) were positively isolated with the MACS system (Miltenyi Biotec). The DX5+ NK cells were suspended in RPMI1640 with 10% FCS and mixed with 51Cr-labeled B16D8 cells at indicated E/T ratios. After 4 h, supernatants were harvested and 51Cr release was measured. Specific lysis was calculated by (specific release − spontaneous release)/(max release − spontaneous release). In some experiments, blood was drawn from the eyes of mice 8 h after polyI:C administration for cytokine measurement.
Test for in vivo NK activation in LN.
5 × 105 WT BMDCs incubated with or without 10 µg/ml polyI:C for 24 h or 5 × 105 IRF-3−/− BMDCs infected with control virus or INAM-expressing lentivirus and allowed to stand for 24 h were injected into the footpads of WT C57BL/6 mice. 48 h later, cells in their inguinal LN were harvested, stained with PE-DX5, and sorted by FACSAria II. RNA was extracted from the DX5-positive cells with TRIzol.
DC therapy.
DC therapy against mice with B16D8 tumor burden was described previously (Akazawa et al., 2007a). C57BL/6 mice (n = 3) were shaved at the flank and injected s.c. with 6 × 105 syngeneic B16D8 melanoma cells (indicated as day 0). For DC therapy, BMDCs were prepared by transfecting control lentivirus or INAM-expressing lentivirus (INAM/pLenti-IRES-hrGFP) and cultured for 24 h. At the time point indicated in the figures, 106 BMDCs were injected s.c. near the tumor. To deplete NK cells in vivo, mice were i.p. injected with hybridoma ascites of anti-NK1.1 mAb (PK136; Akazawa et al., 2007a). Tumor volumes were measured using a caliper every 1 or 2 d. Tumor volume was calculated using the formula: tumor volume (cm3) = (long diameter) × (short diameter) × (short diameter) × 0.4.
Statistical analysis.
Statistical analyses were made with the Student’s t test. The p-value of significant differences is reported.
Online supplemental material.
TICAM-1–inducible genes encoding putative membrane proteins relevant for this study are summarized in Table S1. Fig. S1 shows KO mice results suggesting that both IPS-1 and TICAM-1 in BMDC participate in polyI:C-driven NK activation. Data presented in Fig. S2 characterizes the in vivo polyI:C response of INAM in LN cells. Figs. S3 and S4 demonstrate the properties of surface-expressed INAM analyzed by immunoprecipitation/blotting and confocal microscopy, respectively. Fig. S5 mentions the cytokine expression and maturation profiles of INAM-overexpressing BMDC. Fig. S6 shows the effect of gene silencing of INAM on the polyI:C-mediated cytokine-inducing profile in BMDC. Two pieces of data presented in Fig. S7 confirm the presence of the INAM protein in INAM lentivirus-transduced BMDCs and NK cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091573/DC1.
Acknowledgments
We thank Drs. T. Akazawa and N. Inoue (Osaka Medical Center for Cancer, Osaka, Japan) for their valuable discussions. Thanks are also due to many discussions by our laboratory members. Particularly, extensive English review by Dr. Hussein H. Aly is gratefully acknowledged.
This project was supported by Grants-in-Aid from the Ministry of Education, Science, and Culture and the Ministry of Health, Labor, and Welfare of Japan, Mitsubishi Foundation, Mochida Foundation, NorthTec Foundation Waxman Foundation, and Yakult Foundation.
The authors declare no financial or commercial conflict of interest.
Footnotes
Abbreviations used:
- BMDC
- BM-derived DC
- IKK
- IκB kinase
- INAM
- IRF-3–dependent NK-activating molecule
- IPS-1
- IFN promoter stimulator 1
- IRF
- IFN regulatory factor
- mDC
- myeloid DC
- PRR
- pattern recognition receptor
- Rae-1
- retinoic acid–inducible gene 1
- TICAM-1
- toll/IL-1 receptor homology domain–containing adaptor molecule 1
- TLR
- Toll-like receptor
References
- Akazawa T., Ebihara T., Okuno M., Okuda Y., Shingai M., Tsujimura K., Takahashi T., Ikawa M., Okabe M., Inoue N., et al. 2007a. Antitumor NK activation induced by the Toll-like receptor 3-TICAM-1 (TRIF) pathway in myeloid dendritic cells. Proc. Natl. Acad. Sci. USA. 104:252–257 10.1073/pnas.0605978104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa T., Shingai M., Sasai M., Ebihara T., Inoue N., Matsumoto M., Seya T. 2007b. Tumor immunotherapy using bone marrow-derived dendritic cells overexpressing Toll-like receptor adaptors. FEBS Lett. 581:3334–3340 10.1016/j.febslet.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Azuma M., Sawahata R., Akao Y., Ebihara T., Yamazaki S., Matsumoto M., Hashimoto M., Fukase K., Fujimoto Y., Seya T. 2010. The peptide sequence of diacyl lipopeptides determines dendritic cell TLR2-mediated NK activation. PLoS One. 5:e12550 10.1371/journal.pone.0012550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L., Schjeide B.M., Hooli B., Mullin K., Hiltunen M., Soininen H., Ingelsson M., Lannfelt L., Blacker D., Tanzi R.E. 2008. No association between CALHM1 and Alzheimer’s disease risk. Cell. 135:993–994, author reply :994–996 10.1016/j.cell.2008.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt C.S., Baratin M., Yi E.C., Kennedy J., Gao Z., Fox B., Haldeman B., Ostrander C.D., Kaifu T., Chabannon C., et al. 2009. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 206:1495–1503 10.1084/jem.20090681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot F., Strowig T., Roberts S.M., Arrey F., Münz C. 2007. NK cell survival mediated through the regulatory synapse with human DCs requires IL-15Ralpha. J. Clin. Invest. 117:3316–3329 10.1172/JCI31751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerwenka A., Lanier L.L. 2001. Natural killer cells, viruses and cancer. Nat. Rev. Immunol. 1:41–49 10.1038/35095564 [DOI] [PubMed] [Google Scholar]
- Cerwenka A., Bakker A.B., McClanahan T., Wagner J., Wu J., Phillips J.H., Lanier L.L. 2000. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 12:721–727 10.1016/S1074-7613(00)80222-8 [DOI] [PubMed] [Google Scholar]
- Cerwenka A., Baron J.L., Lanier L.L. 2001. Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc. Natl. Acad. Sci. USA. 98:11521–11526 10.1073/pnas.201238598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreses-Werringloer U., Lambert J.C., Vingtdeux V., Zhao H., Vais H., Siebert A., Jain A., Koppel J., Rovelet-Lecrux A., Hannequin D., et al. 2008. A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer’s disease risk. Cell. 133:1149–1161 10.1016/j.cell.2008.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T., Masuda H., Akazawa T., Shingai M., Kikuta H., Ariga T., Matsumoto M., Seya T. 2007. Induction of NKG2D ligands on human dendritic cells by TLR ligand stimulation and RNA virus infection. Int. Immunol. 19:1145–1155 10.1093/intimm/dxm073 [DOI] [PubMed] [Google Scholar]
- Fernandez N.C., Lozier A., Flament C., Ricciardi-Castagnoli P., Bellet D., Suter M., Perricaudet M., Tursz T., Maraskovsky E., Zitvogel L. 1999. Dendritic cells directly trigger NK cell functions: cross-talk relevant in innate anti-tumor immune responses in vivo. Nat. Med. 5:405–411 10.1038/7403 [DOI] [PubMed] [Google Scholar]
- Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491–496 10.1038/ni921 [DOI] [PubMed] [Google Scholar]
- Gerosa F., Baldani-Guerra B., Nisii C., Marchesini V., Carra G., Trinchieri G. 2002. Reciprocal activating interaction between natural killer cells and dendritic cells. J. Exp. Med. 195:327–333 10.1084/jem.20010938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman J.A., Ogasawara K., Lanier L.L. 2004. Cutting edge: Toll-like receptor signaling in macrophages induces ligands for the NKG2D receptor. J. Immunol. 172:2001–2005 [DOI] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Hornung V., Rothenfusser S., Britsch S., Krug A., Jahrsdörfer B., Giese T., Endres S., Hartmann G. 2002. Quantitative expression of toll-like receptor 1-10 mRNA in cellular subsets of human peripheral blood mononuclear cells and sensitivity to CpG oligodeoxynucleotides. J. Immunol. 168:4531–4537 [DOI] [PubMed] [Google Scholar]
- Huntington N.D., Legrand N., Alves N.L., Jaron B., Weijer K., Plet A., Corcuff E., Mortier E., Jacques Y., Spits H., Di Santo J.P. 2009. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 206:25–34 10.1084/jem.20082013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Medzhitov R. 2004. Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5:987–995 10.1038/ni1112 [DOI] [PubMed] [Google Scholar]
- Kalinski P., Mailliard R.B., Giermasz A., Zeh H.J., Basse P., Bartlett D.L., Kirkwood J.M., Lotze M.T., Herberman R.B. 2005. Natural killer-dendritic cell cross-talk in cancer immunotherapy. Expert Opin. Biol. Ther. 5:1303–1315 10.1517/14712598.5.10.1303 [DOI] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- Kubin M.Z., Parshley D.L., Din W., Waugh J.Y., Davis-Smith T., Smith C.A., Macduff B.M., Armitage R.J., Chin W., Cassiano L., et al. 1999. Molecular cloning and biological characterization of NK cell activation-inducing ligand, a counterstructure for CD48. Eur. J. Immunol. 29:3466–3477 [DOI] [PubMed] [Google Scholar]
- Lee A.E., Rogers L.A., Longcroft J.M., Jeffery R.E. 1990. Reduction of metastasis in a murine mammary tumour model by heparin and polyinosinic-polycytidylic acid. Clin. Exp. Metastasis. 8:165–171 10.1007/BF00117789 [DOI] [PubMed] [Google Scholar]
- Levy S., Shoham T. 2005. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5:136–148 10.1038/nri1548 [DOI] [PubMed] [Google Scholar]
- Lucas M., Schachterle W., Oberle K., Aichele P., Diefenbach A. 2007. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 26:503–517 10.1016/j.immuni.2007.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda H., Saeki Y., Nomura M., Shida K., Matsumoto M., Ui M., Lanier L.L., Seya T. 2002. High levels of RAE-1 isoforms on mouse tumor cell lines assessed by anti-“pan” RAE-1 antibody confer tumor susceptibility to NK cells. Biochem. Biophys. Res. Commun. 290:140–145 10.1006/bbrc.2001.6165 [DOI] [PubMed] [Google Scholar]
- Matsumoto M., Seya T. 2008. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv. Rev. 60:805–812 10.1016/j.addr.2007.11.005 [DOI] [PubMed] [Google Scholar]
- McCartney S., Vermi W., Gilfillan S., Cella M., Murphy T.L., Schreiber R.D., Murphy K.M., Colonna M. 2009. Distinct and complementary functions of MDA5 and TLR3 in poly(I:C)-mediated activation of mouse NK cells. J. Exp. Med. 206:2967–2976 10.1084/jem.20091181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Janeway C.A., Jr 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 91:295–298 10.1016/S0092-8674(00)80412-2 [DOI] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 437:1167–1172 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Miyake T., Kumagai Y., Kato H., Guo Z., Matsushita K., Satoh T., Kawagoe T., Kumar H., Jang M.H., Kawai T., et al. 2009. Poly I:C-induced activation of NK cells by CD8 alpha+ dendritic cells via the IPS-1 and TRIF-dependent pathways. J. Immunol. 183:2522–2528 10.4049/jimmunol.0901500 [DOI] [PubMed] [Google Scholar]
- Mukai M., Imamura F., Ayaki M., Shinkai K., Iwasaki T., Murakami-Murofushi K., Murofushi H., Kobayashi S., Yamamoto T., Nakamura H., Akedo H. 1999. Inhibition of tumor invasion and metastasis by a novel lysophosphatidic acid (cyclic LPA). Int. J. Cancer. 81:918–922 [DOI] [PubMed] [Google Scholar]
- Newman K.C., Riley E.M. 2007. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat. Rev. Immunol. 7:279–291 10.1038/nri2057 [DOI] [PubMed] [Google Scholar]
- Nomura M., Zou Z., Joh T., Takihara Y., Matsuda Y., Shimada K. 1996. Genomic structures and characterization of Rae1 family members encoding GPI-anchored cell surface proteins and expressed predominantly in embryonic mouse brain. J. Biochem. 120:987–995 [DOI] [PubMed] [Google Scholar]
- Ohteki T., Tada H., Ishida K., Sato T., Maki C., Yamada T., Hamuro J., Koyasu S. 2006. Essential roles of DC-derived IL-15 as a mediator of inflammatory responses in vivo. J. Exp. Med. 203:2329–2338 10.1084/jem.20061297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. 2003a. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 4:161–167 10.1038/ni886 [DOI] [PubMed] [Google Scholar]
- Oshiumi H., Sasai M., Shida K., Fujita T., Matsumoto M., Seya T. 2003b. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J. Biol. Chem. 278:49751–49762 10.1074/jbc.M305820200 [DOI] [PubMed] [Google Scholar]
- Sasai M., Shingai M., Funami K., Yoneyama M., Fujita T., Matsumoto M., Seya T. 2006. NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type I IFN induction. J. Immunol. 177:8676–8683 [DOI] [PubMed] [Google Scholar]
- Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 13:539–548 10.1016/S1074-7613(00)00053-4 [DOI] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 122:669–682 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Seya T., Matsumoto M. 2009. The extrinsic RNA-sensing pathway for adjuvant immunotherapy of cancer. Cancer Immunol. Immunother. 58:1175–1184 10.1007/s00262-008-0652-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivori S., Falco M., Della Chiesa M., Carlomagno S., Vitale M., Moretta L., Moretta A. 2004. CpG and double-stranded RNA trigger human NK cells by Toll-like receptors: induction of cytokine release and cytotoxicity against tumors and dendritic cells. Proc. Natl. Acad. Sci. USA. 101:10116–10121 10.1073/pnas.0403744101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Mori Y., Ishii H., Akedo H. 1988. Enhancement of metastatic capacity of fibroblast-tumor cell interaction in mice. Cancer Res. 48:1456–1459 [PubMed] [Google Scholar]
- Tsuji S., Uehori J., Matsumoto M., Suzuki Y., Matsuhisa A., Toyoshima K., Seya T. 2001. Human intelectin is a novel soluble lectin that recognizes galactofuranose in carbohydrate chains of bacterial cell wall. J. Biol. Chem. 276:23456–23463 10.1074/jbc.M103162200 [DOI] [PubMed] [Google Scholar]
- Vivier E., Tomasello E., Baratin M., Walzer T., Ugolini S. 2008. Functions of natural killer cells. Nat. Immunol. 9:503–510 10.1038/ni1582 [DOI] [PubMed] [Google Scholar]
- Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., Shu H.B. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 19:727–740 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. 2003a. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Uematsu S., Hoshino K., Kaisho T., Takeuchi O., Takeda K., Akira S. 2003b. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 4:1144–1150 10.1038/ni986 [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- Zanoni I., Foti M., Ricciardi-Castagnoli P., Granucci F. 2005. TLR-dependent activation stimuli associated with Th1 responses confer NK cell stimulatory capacity to mouse dendritic cells. J. Immunol. 175:286–292 [DOI] [PubMed] [Google Scholar]
- Zou Z., Nomura M., Takihara Y., Yasunaga T., Shimada K. 1996. Isolation and characterization of retinoic acid-inducible cDNA clones in F9 cells: a novel cDNA family encodes cell surface proteins sharing partial homology with MHC class I molecules. J. Biochem. 119:319–328 [DOI] [PubMed] [Google Scholar]