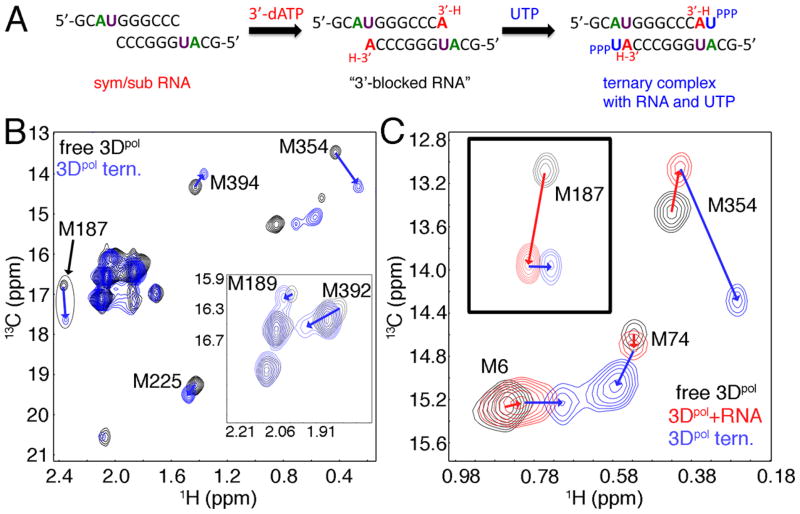
Figure 3.
Conformational changes upon binding RNA and nucleotide to WT 3Dpol. A. Strategy to generate 3Dpol complexes bound with RNA and nucleotide. Addition of 3′-dATP to the 3Dpol/RNA reaction mixture will result in nucleotide incorporation into the sym/sub RNA, but will prevent additional phosphodiester bond formation with subsequent addition of UTP nucleotide. This will essentially “trap” the enzyme in a ternary complex bound with 3′-blocked RNA and UTP. B. 1H-13C HSQC comparison of WT 3Dpol in the unliganded state (black) and in the ternary complex with 3′-blocked RNA and UTP (blue). C. Closeup of the Met6, Met74 and Met354 resonances showing a comparison between unliganded enzyme (black), enzyme bound with sym/sub RNA (before 3′-dATP addition) (red) and enzyme bound with 3′-blocked RNA and UTP (blue). 3Dpol (245 μM) was in 2H2O-based 10 mM HEPES pH 8.0, 200 mM NaCl, 5 mM MgCl2, 10 μM ZnCl2 and 0.02% NaN3. Sym/sub RNA and UTP were at concentrations of 490 μM (duplex concentration) and 4 mM, respectively. Spectra were acquired at 293 K.