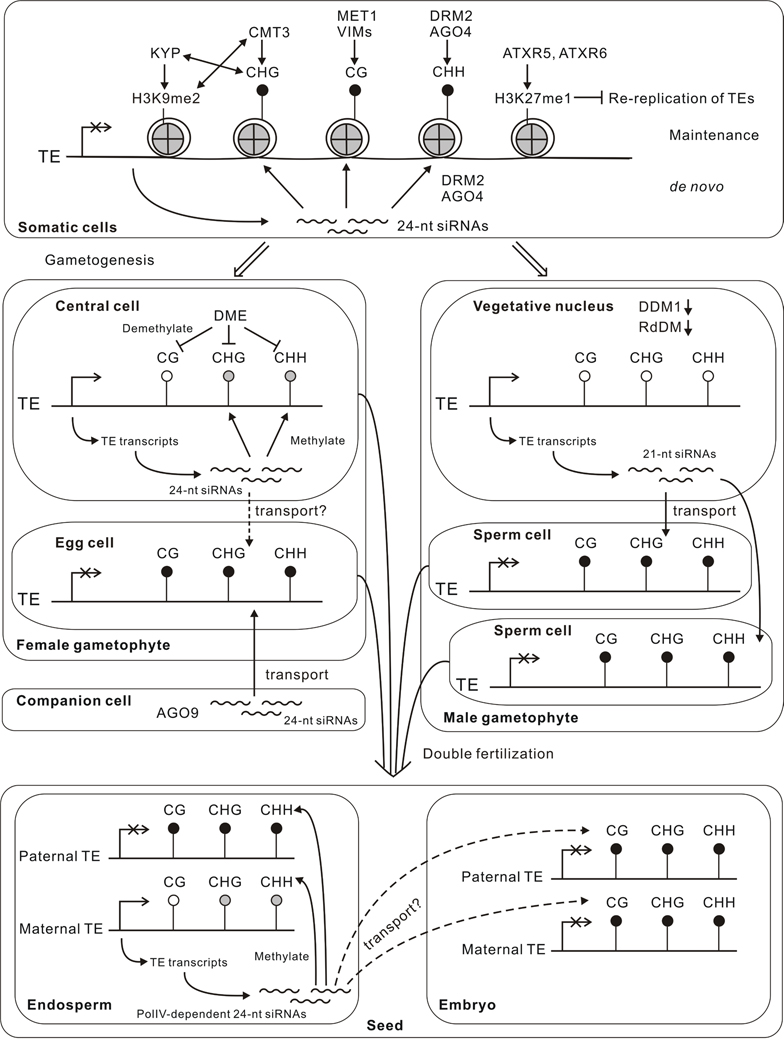
Fig. 1.
Model of epigenetic silencing dynamics during Arabidopsis life cycle. In somatic cells, three different mechanisms are responsible for repressing transcription from transposable element (TE), DNA methylation (in all three sequence contexts), histone H3K9 dimethylation (H3K9me2), and histone H3K27 monomethylation (H3K27me1). Methyltransferases and proteins regulating these epigenetic marks are shown in the diagram. See text for details. In the female gametophyte, the central cell is demethylated by DME, which leads to TE activation and upregulation of RdDM. The siRNAs produced from TEs not only direct non-CG methylation in the central cell, but also might travel to egg cell and enhance the silencing of TEs there. In addition, AGO9-associated siRNAs produced in somatic companion cells also contribute to the silencing of TEs in the egg cell. In the male gametophyte, the vegetative nucleus does not express DDM1 and has reduced RdDM, which leads to TE activation and mobilization. A new class of 21-nt siRNAs is produced from TEs in the vegetative nucleus that travels to sperm cells to reinforcing TE silencing. After double fertilization, maternal TEs in the endosperm stay activated and produce PolIV-dependent siRNAs, which could function to silence the paternal TEs in the endosperm. The methylation levels in the embryo are elevated, possibly due to the siRNA signals transmitted from the endosperm. Different shadings indicate the level of DNA methylation, with black being the high, gray being medium, and white being low.