Abstract
Background
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) is a global healthcare problem. The purpose of this study was to characterize CA-MRSA clones and their distribution in Kuwait hospitals.
Methods
In total, 135 CA-MRSA isolates, carrying the SCCmec IV or V genetic elements, isolated in eight hospitals were characterized using antibiogram, pulsed-field gel electrophoresis, multilocus sequence typing, and carriage of genes for Panton-Valentine Leukocidin (PVL), capsular polysaccharides types (cap) 5 and 8, accessory genes regulators (agr), Staphylococcal enterotoxins (SE) and toxic shock syndrome toxin 1 (tst).
Results
They were susceptible to vancomycin, teicoplanin and linezolid but resistant to kanamycin (62%), fusidic acid (42.2%), tetracycline (39.3%), erythromycin and clindamycin (21.5%), gentamicin (5.9%), streptomycin (6.7%), trimethoprim (5.9%), mupirocin (6.6%) and cadmium acetate (82.2%). They consisted of 10 pulsotypes with the majority belonging to PFGE type I (51.1%), type II (22.2%), type IV (13.3%) and type III (3.7%). They belonged to 10 sequence types (ST) comprising ST80 (51.1%), ST30 (22.2%), ST5 (14.1%), ST1 (4.45), ST6 (3.7%), ST88 (1.5%), ST834 (1.5%), ST8 (0.7%), ST46 (0.7%) and ST950 (0.7%). Genes for PVL, cap 8, cap 5 and agr III, agr I and agr II were detected in 61.5%, 77.3%, 20.7% and 62.2%, 17% and 8.1% of the isolates respectively. Nine (6.7%) isolates contained tst while 103 isolates were positive for SE genes with sei (63.0%), seg (41.5%) and sed (29.6%) as the common SE genes.
Conclusions
ST80-SCCmecIV was the most common CA-MRSA clone in Kuwait hospitals presenting new challenges for infection control.
Background
Following their initial reports from patients in remote communities with no access to healthcare facilities [1-3], Community-associated methicillin resistant S. aureus (CA-MRSA) has now been reported in many countries and has become a global public health problem [4,5]. CA-MRSA carry the smaller staphylococcal chromosome cassette mec (SCCmec) types IV and V whereas the larger SCCmec types I, II and III are present in MRSA traditionally associated with infections in healthcare facilities [4,5]. Initially, CA-MRSA were generally more susceptible to non beta-lactam antibiotics than healthcare associated MRSA [1,4,5]. However, some CA-MRSA strains have now acquired multidrug resistance [6-9].
The transmission of CA-MRSA was restricted to particular populations with distinctive risk factors such as injections drug users and aboriginal communities [2,4,5]. However, since the 1990's CA-MRSA has become widespread and have been introduced from the community into healthcare facilities where, in some instances, they have become the dominant MRSA clones [10,11]. They have demonstrated fitness to survive in healthcare facilities. Consequently, some epidemic CA-MRSA clones have emerged in several locations. These include the USA300 clone which is dominant in USA and North America [12], the ST30 (Oceanic clone) [13] and the European ST80 clone [14-16].
CA-MRSA was first isolated in small numbers in Kuwait hospitals in 2001 [17] when it constituted 1.8% of MRSA obtained from patients in seven hospitals. Molecular typing of these isolates revealed ST30 and ST80 as the common CA-MRSA clones [17]. Since then, the prevalence of CA-MRSA has increased substantially and by 2005, 17% of all MRSA isolated in Kuwait hospitals were CA-MRSA [18]. However, there was no information on the clonal composition of these strains. The objective of this study was to characterize CA-MRSA obtained in Kuwait hospitals between 2005 and 2006 using SCCmec typing, PFGE and multi-locus sequence typing to ascertain their clonal composition and prevalence.
Materials and methods
MRSA isolates
From January 2005 to December 2006, the MRSA Reference Laboratory, located at the Department of Microbiology, Faculty of Medicine, Kuwait University, Kuwait, received 889 MRSA for typing. One hundred and thirty five of these isolates were classified as CA-MRSA based on their carriage of SCCmec IV or V genetic elements and urease production. The 135 isolates originated from 131 patients located in eight hospitals. The isolates were collected from Mubarak (MBH, 18 isolates), Amiri (AMH,31 isolates), Al-Sabah (ASH,29 isolates), Farwania (FAH; 12 isolates), Ibn Sina (ISH; 16 isolates), Al-Razi (ARH; 9 isolates), Jahra (JH; 19 isolates) and the Maternity hospital (MAT;1 isolate). from wound swabs (37 isolates), abscesses (18 isolates), nasal swabs (18 isolates), ear swabs (9 isolates), blood samples (5 isolates), groin (5 isolates), throat swabs (5 isolates), urine samples (4 isolates), skin swabs (3 isolates), tracheal aspirates (3 isolates), high vaginal swabs (2 isolates), sputum (2 isolates) and eye swab (1 isolate). The source for 20 isolates was not provided. The four isolates from the same patient were obtained from four different sites and were all resistant to mupirocin [9]. Isolates were identified at the diagnostic laboratories and confirmed as CA-MRSA at the MRSA Reference Laboratory using SCCmec typing and urease production. Pure cultures were preserved in 40% (v/v) glycerol in brain heart infusion broth (BHIB) at -20°C. Isolates were recovered in BHIB, incubated at 37 °C, and sub-cultured on brain heart infusion agar plates before testing.
Susceptibility to antibacterial agents
Susceptibility testing to antibacterial agents was performed by the disk diffusion method [19] using oxacillin, benzyl penicillin, cefoxitin, kanamycin, gentamicin, erythromycin, clindamycin, chloramphenicol, tetracycline, trimethoprim, fusidic acid, rifampicin, ciprofloxacin, and linezolid. Susceptibility to fusidic acid was interpreted following guidelines by the British Society for Antimicrobial Chemotherapy [20]. Disks containing (values per disk): cadmium acetate (50 μg), propamidine isethionate (100 μg), mercuric chloride (109 μg) were prepared in the laboratory. The results were interpreted as reported previously [20]. Disk susceptibility testing for mupirocin was performed with disks containing 5 μg and 200 μg of mupirocin (Oxoid, Basingstoke, UK). Growth to the edge of both the 5 μg and the 200 μg disks indicated high level resistance. Growth to edge of the 5 μg disk alone defined low level resistance [21]. High-level mupirocin resistance was confirmed by MIC determination and the detection of mupA gene by PCR as described previously [9]. The MIC for oxacillin, mupirocin, vancomycin and teicoplanin were determined with Etest strips (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. The Macro method, using an inoculum equivalent to 2× McFarland standard on brain heart infusion agar, was used to determine MIC for vancomycin and teicoplanin [22]. S. aureus strain ATCC25923 was used as quality control strain for susceptibility testing. Methicillin resistance was confirmed by detecting PBP 2a using a rapid latex agglutination kit (Denka-Seiken, Japan) according to the manufacturer's instructions.
Urease production
Urease production was detected on Christensen's urea agar slope after incubation at 35°C for 48 hours.
SCCmec and ccr typing
Staphylococcal cassette chromosome mec (SCCmec) typing and subtyping were performed as described previously [23]. The cassette chromosome recombinase (ccr complex) types was determined using primers and methods described previously [24]. Strain COL (SCCmec type 1, ccr 2); XU642 (EMRSA-16, SCCmec type II, ccr 2); WBG525 (EMRSA-1, SCCmec type III), and K1814 (SCCmec type IV) and WBG 8318(SCCmec type V) as controls.
Pulsed-field gel electrophoresis (PFGE)
Genomic DNA was prepared in agarose plugs as described previously [25]. DNA was digested with 40 U Sma I- (Biolabs, New England, USA) for 4 hr at 25°C. Counter-clamped homogenous electric field (CHEF) electrophoresis of digested chromosomal DNA was performed in 1.2% agarose (pulse-field certified agarose, BioRad, USA) at 6 V/cm for 21 h at 14°C, with pulse times of 5 s-40 s using a CHEF-DRIII system (BioRad). Sma I digested chromosomal DNA of S. aureus strain NCTC8325 was used as a molecular size marker. After electrophoresis, the gel was stained with ethidium bromide 0.5 μg/ml and photographed under ultraviolet illumination. The chromosomal patterns were examined visually and similarities of the PFGE patterns were assigned using criteria published previously [26].
Multilocus sequence typing (MLST)
Multilocus sequence typing (MLST) was performed by following previously published protocols [27]. The sequences of the seven house keeping genes (arc, aroE, glpF, gmk, pta, tpi and yqiL) were compared to existing sequences in the MLST database http://www.mlst.net for assignment of allelic numbers. Sequence types (ST) were assigned according to their allelic profiles.
Detection of virulence genes
DNA was extracted from S. aureus for PCR following a protocol described previously [25]. PCR assays were used to investigate the carriage of genes for accessory gene regulator (agr) types I, II, III and IV using the primers and conditions published previously [28]. The LukS-PV-lukF-PV gene which expresses Panton-Valentine leucocidin (PVL) was amplified as described previously [15]. The amplification of genes for capsular polysaccharide (cap) types 5 and 8 were performed using primers and conditions described previously [28]. Genes for the staphylococcal enterotoxins, sea, seb, see, sed, sei, seh and toxic shock syndrome gene, tst were amplified in multiplex PCR as described previously [25]. All PCR products were analysed by agarose gel electrophoresis on 1.5% agarose gels [25].
Statistics
Differences in the distribution of CA-MRSA clones were assessed by Chi square test. A P-value of ≤ 0.05 was considered significant.
Results
Staphylococcal cassette chromosome mec (SCCmec) and ccr typing
The 135 MRSA isolates included in this study produced urease and belonged to SCCmec type IV or type V. Urease positivity was used to differentiate CA-MRSA carrying the SCCmec IV genetic element from the healthcare-associated epidemic MRSA-15 (EMRSA-15) which also carries the SCCmec-IV genotype but does not produce urease [29]. In total, 102 (75.6%) of the isolates carried SCCmec type IV, 11 (8.1%) carried SCCmec type IVa, 10 (7.4%) carried SCCmec type IVc and 12 (8.9%) the SCCmec type V genetic element. A total of 123 (91.1%) isolates consisting of the SCCmec type IV, SCCmec IVa and SCCmec IVc isolates belonged to ccr type 2 while the 12 SCCmec type V isolates belonged to ccr type 5.
Antibacterial resistance of CA-MRSA isolates
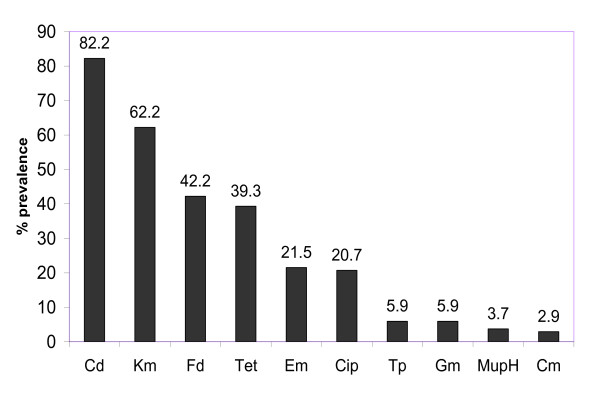
All isolates were susceptible to vancomycin MIC: ≤ 2 μg/ml), teicoplanin MIC: ≤ 2 μg/ml), rifampicin and linezolid but were resistant to oxacillin (MIC 8- > 256 μg/ml) and the antibacterial agents summarized in Figure 1. There was a high prevalence of resistance to cadmium acetate (82.2%), kanamycin (62.2%), fusidic acid (42.2%), tetracycline (39.3%), erythromycin and clindamycin (21.5%) and ciprofloxacin (20.7%). Sixty-eight (50.4%) of the 135 CA-MRSA isolates were multidrug resistant (i.e. resistant to more than three classes of non-beta lactam antibiotics tested).
Figure 1.
Antimicrobial resistance of CA-MRSA isolates. Abbreviations: Cd, cadmium acetate; Kan, kanamycin; Fd, fusidic acid; Tet, tetracycline; Erm, erythromycin; Cip, ciprofloxacin; Tp, trimethoprim; Gen, gentamicin; mupH, high-level mupirocin; Chl, chloramphenicol.
Molecular typing of CA-MRSA isolates
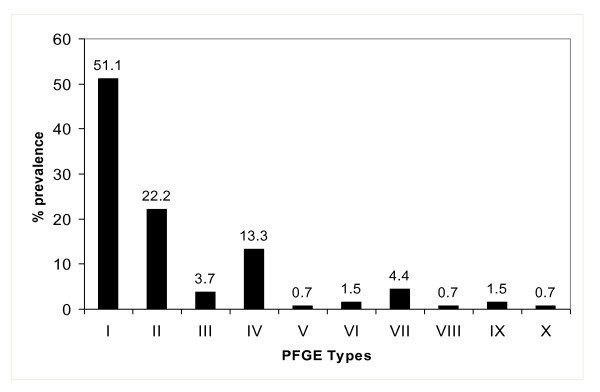
Figure 2 shows the distribution of the 10 PFGE types detected among the 135 isolates. The majority belonged to PFGE type I or its five subtypes (51.1%), PFGE type II and its two subtypes (23.7%), PFGE type IV and subtype IVa (13.3%). The other PFGE types occurred sporadically.
Figure 2.
Distribution of PFGE types among CA-MRSA isolates.
MLST was performed on 28 isolates representing each of the PFGE subtypes to determine their sequence types (ST). MLST results showed extensive agreement with PFGE except for ST5 that was detected in PFGE types IV and IX. The isolate belonging to PFGE type IX was ST5 while the isolate belonging to subtype IXa was ST950 (a SLV of ST5 at tpi). Based on the distribution of sequence types according to their PFGE patterns, 69 (51.1%) of the 135 isolates were ST80. This was followed by ST30 (22.2%), ST5 (14.1%), ST1 (4.4%), ST6 (3.7%), ST88 (1.5%), ST834 (1.5%), ST8 (0.7%), ST46 (0.7%) and ST950 (0.7%). As shown in Table 1, the ST80 clone was present in seven of the eight hospitals. ST30 and ST5 clones were detected in six hospitals while ST1 clone was present in five hospitals. The other clones occurred sporadically. When compared with the distribution of CA-MRSA clones prevalent in Kuwait hospitals in 2001-2003 [17], the proportion of ST80 isolates increased from 38.5% in 2001-2003 [17] to 51.1% (P = 0.29), while the proportion of ST30 isolates decreased from 30.8% in 2001-2003 to 23.7% (P = 0.45)
Table 1.
Distribution of CA-MRSA clones in Kuwait hospitals
| ST | AMH | ARH | ASH | FAH | MBH | JH | ISH | MAT | Total | % |
|---|---|---|---|---|---|---|---|---|---|---|
| 80 | 12 | 4 | 8 | 10 | 14 | 11 | 10 | - | 69 | 51.1 |
| 30 | 10 | - | 11 | - | 2 | 3 | 3 | 1 | 30 | 22.2 |
| 5 | 3 | 4 | 7 | - | 1 | 3 | 1 | - | 19 | 14 |
| 1 | 1 | - | 1 | 2 | 1 | - | 1 | - | 6 | 4.4 |
| 6 | 1 | 2 | - | - | 2 | - | - | - | 5 | 3.7 |
| 88 | - | - | - | 2 | - | - | - | - | 2 | 1.5 |
| 8 | 1 | - | - | - | - | - | - | - | 1 | 0.7 |
| 46 | 1 | - | - | - | - | - | - | - | 1 | 0.7 |
| 950 | 1 | - | - | - | - | - | - | - | 1 | 0.7 |
| 834 | - | - | - | - | - | - | 1 | - | 1 | 0.7 |
| Total | 30 | 10 | 27 | 14 | 20 | 17 | 16 | 1 | 135 | |
Detection of virulence-associated genes
None of the isolates was positive for agr type IV. However, they contained genes for agr III (62.2%), agr I (17%), agr II (8.1%), capsular polysaccharides types 8 (77.3%) and type 5 (20.7%). Three (2.2%) isolates yielded negative result for both cap 5 and cap 8. The PVL gene was detected in 61.5% of the isolates.
In total, 103 (76.3%) isolates yielded positive results for sea (10.4%), seb (3.7%), sec (4.4%), sed (29.6%), seg (41.5%), seh (1.5%), sei (63.0%) and tst (6.7%). None of the isolates was positive for see. Twenty-seven (26.2%), 52 (50.5%), 17 (16.5%) and seven (6.8%), of the 103 isolates were positive for single, two, three and four SE genes respectively. Twenty-three isolates contained seg and sei while 20 isolates contained sed and sei.
Diversity of ST80-CA-MRSA isolates
Table 2 summarizes the phenotypic and genotypic characteristics of the ST80 isolates. The 69 ST80 isolates had related but not identical PFGE patterns. Although 39 of the isolates had identical PFGE patterns (PFGE type I), the remaining 30 isolates were grouped into five subtypes: Ia (2 isolates), Ib (10 isolates), Ic (1 isolate), Id (2 isolates and Ie (15 isolates).
Table 2.
Characteristics of ST80 CA-MRSA isolates.
| Virulence-associated genes (# of positives) | ||||||
|---|---|---|---|---|---|---|
| # of isolates | PFGE | SCCmec | Antibiogram | PVL | SE | tst |
| 4 | I | IV | Cd, Kan, Tet, Fd | + (2) | sei,(2) sed (2)- | - |
| 2 | Ib | IV | Cd, Kan, Tet, Fd | - | sei (1), sed (1), seg (1) | - |
| 1 | Id | IV | Cd, Kan, Tet, Fd | + | - | - |
| 2 | Ie | IV | Cd, Kan, Tet, Fd | + | - | - |
| 3 | I | IV | Cd, Kan, Tet, Fd, Cip | + (2) | sei (2), sed (1) | - |
| 2 | Ie | IV | Cd, Kan, Tet, Fd, Cip | + (2) | sei (2), sed (2) | - |
| 1 | I | Iva | Cd, Kan, Tet, Fd, Cip, Gen | + | sei, seg , | - |
| 4 | Ib | IV | Cd, Kan, Tet, Fd, Mup | + | seg (4) | - |
| 1 | I | IV | Cd, Kan, Tet, Fd, Tp | + | seg | - |
| 1 | I | IV | Cd, Kan, Tet, Fd, Erm | + | sei | - |
| 1 | Ie | IV | Cd, Kan, Tet, Fd, Gen | - | seg | - |
| 2 | I | IV | Cd, Kan, Fd, Erm, Cip | + (1) | sei (1), seg (1) | - |
| 1 | I | IV | Cd, Kan, Fd, Erm, Cip, Chl | - | sed | - |
| 1 | I | IV | Cd, Kan, Fd, Cip | - | sei | - |
| 1 | Ib | IV | Cd, Kan, Fd, Cip | + | sei, seg | - |
| 1 | I | IV | Cd, Kan, Fd, Gen | + | seg | - |
| 1 | Ib | IV | Cd, Kan, Fd, Gen | - | - | - |
| 1 | I | IV | Cd, Kan, Fd | + | sei | - |
| 1 | Ib | IV | Cd, Kan, Fd | + | sei,, sed | + |
| 3 | I | IV | Cd, Kan | + (3) | sei,(2) sed (2) | - |
| 1 | Ia | IV | Cd, Kan | + | sei | - |
| 1 | Ie | IV | Cd, Kan, Erm, Tet | - | - | - |
| 1 | Ie | IV | Cd, Kan, Erm, Cip | - | sei, sed | - |
| 2 | I | IV | Cd, Kan, Erm, Cip | + (1) | Sei (1), seg (1) | - |
| 1 | Ia | IV | Cd, Kan, Erm, Cip, Chl | + | - | - |
| 1 | I | IV | Cd, Kan, Cip, Tp, Gen | + | sei | - |
| 7 | I | IV | Cd | + | sei (2), sed (1), seg (1) | |
| 1 | Ie | IV | Cd | + | - | - |
| 2 | Ib | IV | Cd | - | sei (2),, seg (2), sec(1) | - |
| 3 | I | IV | Cd, Erm | + | sei (3), seg (2), Sed (1) | - |
| 1 | I | IV | Cd, Fd | + | - | - |
| 2 | I | IV | Cd, Tet | + | Sei (1), seg(2) | - |
| 1 | Ic | IV | Cd, Tet, Fd, Chl, Tp, Cip, Erm | - | sei, sed, seg | - |
| 1 | I | IV | Erm, Cip | + | sei, seb, seg | - |
| 1 | Ie | IVc | Kan, Tet, Fd | + | - | - |
| 1 | Ie | IV | Kan, Tet, Fd | + | sei, sed | - |
| 1 | Ib | IV | Kan | + | sei, sed | - |
| 2 | Ie | IV | Kan | + (2) | sei (1), sed(1) | - |
| 1 | Ie | IV | Kan, Erm | + | - | - |
| 1 | Ie | IV | Tet | - | sei, sed, sea | - |
| 1 | Id | IV | - | + | sea | - |
| 1 | I | IV | - | + | sei | + |
| 1 | Ie | IV | - | - | - | - |
Abbreviations:Cd, cadmium acetate; Cip, ciprofloxacin; Chl. chloramphenicol; Erm, erythromycin; Fd, fusidic acid; Gen, gentamicin; Kan, kanamycin; Tet, tetracycline; Tp, trimethoprim.
The ST80 isolates expressed low oxacillin MIC (MIC 8-64 μg/ml). They also expressed diverse resistance phenotypes. In total, 23 of the 69 isolates were resistant to kanamycin, tetracycline and fusidic acid, that are traditionally associated with ST80-SCCmec-IV isolates (17, 28). Of these 23 isolates, 15 expressed additional resistance to one or more of ciprofloxacin, gentamicin, erythromycin, trimethoprim, mupirocin and chloramphenicol. Furthermore, some of the isolates were susceptible to kanamycin, tetracycline and fusidic acid that usually characterize ST80-SCCmec-IV isolates. Ten isolates were resistant to cadmium acetate but were susceptible to all of the non beta-lactam agents tested. Forty one (59.4%) of the 69 isolates were multiresistant (resistance to more than three antibiotic classes).
All ST80 isolates carried genes for capsular polysaccharide types 8 and agr III but varied in the carriage of other virulence-associated genes. As presented in Table 2, 58 (84.0%) and 49 (71.0%) of the 69 isolates were positive for PVL and SE genes respectively. Fifteen isolates were positive for single SE gene while 30 were positive for two or more SE genes. The majority of the isolates were positive for sei (77.6%) and seg (36.7%). Nineteen (27.9%) isolates were positive for sed and sei, and ten (14.7%) were positive for seg and sei. Two isolates were positive for tst.
Discussion
This study characterized ten different CA-MRSA clones in Kuwait hospitals with ST80 (51.1%) and ST30 (22.2%) as the major CA-MRSA clones similar to the composition of CA-MRSA isolates circulating in Kuwait hospitals in 2001-2003 [17]. However, results of this study differ from the previous study in three aspects. The 2001-2003 isolates consisted of five CA-MRSA clones whereas the present study has identified ten CA-MRSA clones. This represents a substantial increase in the number of CA-MRSA clones and confirms their expansion in Kuwait hospitals. Secondly, whereas the proportion of ST80 isolates increased from 38.5% in 2001-2003 [17] to 51.1% in the present study, the proportion of ST30 isolates decreased from 30.8% in 2001-2003 to 22.2%. However, these changes were not statistically significant. The proportion of ST5 isolates has also increased from 7.7% in 2001-2003 to 14.1%. Thirdly, new clones, ST46, ST88, ST1, ST834 and ST950, appeared for the first time in Kuwait in this study. In contrast, strains belonging to ST361 and ST728 that were present in 2001-2003 were absent in this study highlighting the evolving nature of CA-MRSA isolates.
There was a high prevalence of resistance to cadmium acetate, kanamycin, fusidic acid, tetracycline, erythromycin, clindamycin and ciprofloxacin with 50.4% of the isolates being multiresistant to non-beta lactam antibiotics. Multiresistant-CA-MRSA isolates have also been reported among CA-MRSA clones isolated in Brazil [7], Japan [8] and USA [12].
We detected genes for PVL in 61.5% of the CA-MRSA isolates distributed among different sequence types as has also been reported in other countries [10,12,30,31]. We also detected agr type III genes in 67.4% of our CA-MRSA which is similar to results of studies in Belgium, France, Japan and Switzerland where the majority of their CA-MRSA isolates belong to agr type III [12,16] and possessed capsular polysaccharide type 8 [31]. However, whereas sei (63%) was the most common SE gene among our CA-MRSA isolates, seg was the most abundant SE gene in MRSA obtained in Ireland [30]. We observed the co presence of seg and sei in our isolates which has also been reported in CAMRSA studied in USA [32]. Furthermore, similar to the observation in CA-MRSA isolates from Ireland [30] and USA [32], none of our CA-MRSA isolates was positive for see. However, in contrast to the low prevalence of tst (6.7%) in this study, Rossney et al., [30] detected tst in 24% of CA-MRSA isolates in Ireland.
The ST80-SCCmec-IV clone has been the dominant CA-MRSA clone in European countries for some time [14,16]. With a prevalence of 51.1% of the CA-MRSA, the ST80 clone was the most common CA-MRSA clone in Kuwait hospitals. However, our ST80 isolates varied phenotypically. Although some were resistant to kanamycin, tetracycline and fusidic acid similar to the resistance pattern of the European ST80 clone [14,16], the results in Table 2 showed that some of our ST80 isolates had varied resistance patterns with some being susceptible to kanamycin, tetracycline and fusidic acid. They were also heterogeneous in their enterotoxin gene profiles. Remarkably, our ST80 isolates contained sed, sei, seg, seb, seh and sea, singly or in combinations. In contrast, studies from the Netherlands [16] detected only seh in their ST80 isolates. The detection of genes for different SEs in this study suggests that ST80 isolates may be acquiring genes for different SEs probably due to the acquisition of toxin-carrying bacteriophages [33]. In addition, 11 (16%) of the 69 ST80 isolates were negative for PVL genes. In a previous study, a small number (5 of 357) of ST80 isolates obtained from Algeria, Switzerland and France also lacked genes for PVL [34]. It is uncertain whether these isolates arose from PVL positive isolates due to the loss of the PVL phage or represents native ST80 backgrounds that had not previously acquired the PVL phage.
The dominance of the ST80 and ST30 CA-MRSA clones in Kuwait hospitals in 2001-2003 was attributed to their importation into the country by returning patients who sought medical care abroad, or expatriate staff from countries where these clones are prevalent [17]. Their presence in seven of the eight hospitals during the current study suggests that local transmission is also occurring. In addition, the diverse phenotypes observed with the ST80 isolates suggest that local variants of ST80 clones are emerging in Kuwait.
Conclusion
This study has shown that the ST80-SCCmec-IV clone was the most common CA-MRSA clone in Kuwait hospitals in 2005-2006. However, other sequence types such as ST5 and ST6 also appeared. Most of the isolates were multi-resistant to antibacterial agents and harbored genes for different virulence factors. The successful expansion of CA-MRSA clones in Kuwait hospitals calls for novel approaches in infection control measures to control their spread.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
EEU designed the study, procured funding, guided the experimental work and wrote the first draft of the paper. ES contributed to the study design, performed experimental work and contributed to writing the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Edet E Udo, Email: EDET@hsc.edu.kw.
Eiman Sarkhoo, Email: boushehri1@hotmail.com.
Acknowledgements
This study was supported by project grant YM 05/06 from Kuwait University Research Administration. E. Sarkhoo was a recipient of a Graduate Student fellowship from the College of Graduate Studies, Kuwait University. Kuwait. We thank Bindu Mathews and Bobby Noronha for technical assistance. This study was presented as a poster at the 14th International Congress on Infectious Diseases (ICID), Miami, Florida, March 9-12, 2010.
References
- Udo EE, Pearman JW, Grubb WB. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:157–165. doi: 10.1016/0195-6701(93)90100-E. [DOI] [PubMed] [Google Scholar]
- Groom AV, Wolsey DH, Naimi TS, Smith K, Johnson S, Boxrud D, Moore KA, Cheek JE. Community-acquired methicillin-resistant Staphylococcus aureus in a rural American Indian community. JAMA. 2001;286:1201–1205. doi: 10.1001/jama.286.10.1201. [DOI] [PubMed] [Google Scholar]
- Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Reverdy ME, Etienne J. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emer Infect Dis. 2003;9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltezou HC, Giamarellou HC. Community - acquired methicillin-resistant Staphylococcus aureus infection. Int J Antimicrob Agents. 2006;27:87–96. doi: 10.1016/j.ijantimicag.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired meticillin-resistant: an emerging threat. Lancet Infect Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- Diep BA, Chambers HH, Graber CJ, Szumowskli JD, Miller LG, Han LL, Chen JH, Lin F, Lin J, Phan TH, Carlton HA, McDougal LK, Tenover FC, Cohen DE, Mayer KH, Sensabaugh GF, Pardeau-Remington F. Emergence of multidrug-resistant, community-associated methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 2008;148:249–257. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- Bonelli RR, Souza RR, Moreira S, dos Santos LCG, Conceicado MS, de Mello SJ, Carballido JM, Rito PN, Vieira VV, Teixeira LA, Sa Figueredo AM. Emergence of multiresistant variants of community-acquired methicillin-resistant Staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, Brazil. Diagn Microbiol Infect Dis. 2009;65:300–305. doi: 10.1016/j.diagmicrobio.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Takizawa Y, Taneike I, Nakagawa S, Oishi T, Nitahara Y, Iwakura N, Ozaki K, Takano M, Nakayama T, Yamamoto T. A Panton-Valentine Leucocidin (PVL)-Positive Community-acquired Methicillin-Resistant Staphylococcus aureus (MRSA) strain, another such strain carrying a multiple-drug resistance plasmid, and other more-typical PVL-negative MRSA strains found in Japan. J Clin Microbiol. 2005;43:3356–3363. doi: 10.1128/JCM.43.7.3356-3363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, Sarkhoo E. Genetic analysis of high-level mupirocin resistance in the ST80 clone of community-associated methicillin-resistant Staphylococcus aureus. J Med Microbiol. 2010;59:193–199. doi: 10.1099/jmm.0.013268-0. [DOI] [PubMed] [Google Scholar]
- Hsu L, Koh Y, Chlebicka NL, Tan T, Krishnan P, Lin RT, Tee N, Barkham T, Koh T. Establishment of ST30 as the Predominant Clonal Type among community-associated Methicillin-Resistant Staphylococcus aureus Isolates in Singapore. J Clin Microbiol. 2006;44:1090–1093. doi: 10.1128/JCM.44.3.1090-1093.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran GL, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- Tenover FC, McDougal LK, Goering RV, Killgore G, Projan SJ, Patel JB, Dunman PM. Characterization of a strain of community - associated methicillin - resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol. 2006;44:108–118. doi: 10.1128/JCM.44.1.108-118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari RP, Cook GM, Lamont I, Lang S, Hefferman H, Smith JMB. Phenotypic and molecular characterization of community occurring Western Samoan phage pattern methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 2002;50:825–831. doi: 10.1093/jac/dkf242. [DOI] [PubMed] [Google Scholar]
- Witte W, Braulke C, Cuny C, Strommenger B, Werner G, Heuck D, Jappe U, Wendt C, Linde HJ, Harmsen D. Emergence of methicillin-resistant Staphylococcus aureus with Panton-Valentine leukocidin in central Europe. Eur J Clin Microbial Infect Dis. 2005;24:1–5. doi: 10.1007/s10096-004-1262-x. [DOI] [PubMed] [Google Scholar]
- Piemont Y, Godail-Gamot F, Bes M, Peter M-OM, Gauduchon V, Vandenesch F, Etienne J. Involvement of Panton-Valentine leukocidin producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- Stam-Bolink EM, Mithoe D, Baas WH, Arends JP, Möller AVM. Spread of a methicillin-resistant Staphylococcus aureus ST80 strain in the community of the northern Netherlands. Eur J Clin Microbiol Infect Dis. 2007;26:723–727. doi: 10.1007/s10096-007-0352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, O'Brien FG, Al-Sweih N, Noronha B, Mathew B, Grubb WB. Genetic lineages of community-associated methicillin-resistant Staphylococcus aureus in Kuwait hospitals. J Clin Microbiol. 2008;46:3514–3516. doi: 10.1128/JCM.00966-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, Al-Sweih N, Dhar R, Dimitrov TS, Mokaddas EM, Johny M, Al-Obaid IA, Gomaa HH, Mobasher LA, Rotimi VO, Al-Asar A. Surveillance of antibacterial resistance in Staphylococcus aureus isolated in Kuwaiti hospitals. Med Princ Pract. 2008;17:71–75. doi: 10.1159/000109594. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility testing. 16th information supplement. Approved standard M100-S16. Clinical and Laboratory Standard Institute, Wayne. PA; 2006. [Google Scholar]
- British Society for Antimicrobial Chemotherapy. BSAC Disc diffusion method for Antimicrobial susceptibility testing. British Society for Antimicrobial Chemotherapy. http://www.bsac.org.uk/Resources/BSAC/version_4_january_2005_final_NH_April_2.pdf [DOI] [PubMed]
- Udo EE, Farook VS, Mokadas EM, Jacob LE, Sanyal SC. Molecular fingerprinting of mupirocin-resistant methicillin-resistant Staphylococcus aureus from a Burns unit. Int J Infect Dis. 1999;3:82–87. doi: 10.1016/S1201-9712(99)90014-0. [DOI] [PubMed] [Google Scholar]
- Etest application sheet. EA003. Staphylococci. 2007. http://www.abbiodisk.com
- Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–5033. doi: 10.1128/JCM.43.10.5026-5033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Ma XX, Takeuchi F, Okuma K, Yuzawa H, Hiramatsu K. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48:2637–2651. doi: 10.1128/AAC.48.7.2637-2651.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE, Al-Mufti S, Albert MJ. The prevalence of antimicrobial resistance and carriage of virulence genes in Staphylococcus aureus isolated from food handlers in Kuwait City restaurants. BMC Res Notes. 2009;2:108. doi: 10.1186/1756-0500-2-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright MC, Day NPJ, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PCL, Lindsay JA. Molecular characterization of the dominant UK methicillin-sensitive Staphylococcus aureus strains, EMRSA-15 and EMRSA-16. J Med Microbiol. 2002;51:516–521. doi: 10.1099/0022-1317-51-6-516. [DOI] [PubMed] [Google Scholar]
- Richardson JF, Reith S. Characterization of a strain of methicillin-resistant Staphylococcus aureus (EMRSA-15) by conventional and molecular methods. J Hosp Infect. 1993;25:45–52. doi: 10.1016/0195-6701(93)90007-M. [DOI] [PubMed] [Google Scholar]
- Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O'Connell B, Coleman DC. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine Leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol. 2007;45:2554–2563. doi: 10.1128/JCM.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier I, Durand G, Bes M, Taylor KL, Lina G, Vandenesch F, Fattom AI, Etienne J. Identification of the Capsular Polysaccharides in Staphylococcus aureus Clinical Isolates by PCR and agglutination tests. J Clin Microbiol. 2007;45:725–729. doi: 10.1128/JCM.01572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney AK, Mediavilla JR, Robiou N, Guh A, Wang X, Gialanella P, Levi MH, Kreiswirth BN, Fries BC. Diverse enterotoxin gene profiles among clonal complexes of Staphylococcus aureus isolates from the Bronx, New York. Appl Environ Microbiol. 2009;75:6839–49. doi: 10.1128/AEM.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita S, Kaneko J, Chiba J, Piedmont Y, Jarraud S, Etienne J, Kamio Y. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. Gene. 2001;268:195–206. doi: 10.1016/S0378-1119(01)00390-0. [DOI] [PubMed] [Google Scholar]
- Tristan A, Bes M, Meugnier H, Lina G, Bozdogan B, Courvalin P, Reverdy M-E, Enright MC, Vandenesch F, Etienne J. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus, 2006. Emerg Infect Dis. 2007;13:594–600. doi: 10.3201/eid1304.061316. [DOI] [PMC free article] [PubMed] [Google Scholar]