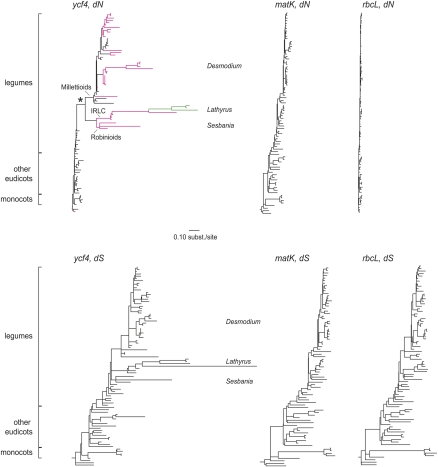
Figure 1.
Synonymous and nonsynonymous divergence in angiosperm chloroplast ycf4 sequences. Shown are dN (upper) and dS (lower) trees resulting from a codon-based likelihood analysis and a constrained topology, rooted using gymnosperm sequences (which are not included in the trees). All trees are drawn to the same scale. The species are in the same order from top to bottom in all trees, to the greatest extent possible, and are named in full in Supplemental Figure S1. Magenta branches in the dN tree for ycf4 indicate those on which the Ycf4 protein length is (or is inferred to have been) ≥200 amino acid residues; green branches indicate lengths ≥300 residues. The asterisk marks the branch (leading to Millettioids, Robinioids, and IRLC) in which rate acceleration is first seen. Trees for chloroplast rbcL and matK genes do not show comparable rate heterogeneity at either synonymous or nonsynonymous sites.