Abstract
Exploring the capabilities of instrumental techniques for discriminating n-3 rich oils derived from animals is a very important though much neglected area that was emphasized more than 100 years ago. In this study the potential of gas chromatography (GC) for discriminating full fatty acid methyl ester (FAME) profiles from fish (cod liver and salmon) and marine mammal (seal and whale) oils is evaluated by means of principal component analysis (PCA). The FAME profiles from plant oils such as rapeseed, linseed and soy oils and seven different brands of n-3 supplements are also used in the discrimination process. The results from the PCA plots can reliably distinguish between plant, n-3 supplements, fish and marine mammal oils. By removing the contribution of the n-3 supplements and plant oils it is possible to discriminate between types of fish and marine animal oils. GC offers a rapid, simple and convenient means of discriminating oils from different species, brands and grades.
Keywords: Marine oil, Discrimination, Gas chromatography, Principal component analysis
Introduction
The importance of developing techniques aiming at detecting adulteration of fish oils has been emphasized since the late nineteenth early twentieth century when a great scarcity of cod liver oil accompanied by famine prices on the market brought about adulteration of genuine cod liver oil with low-grade shark oil [1, 2]. An overview of the different instrumental techniques used for the analysis of plant, fish and marine mammal oils in studies where the terms discrimination, adulteration, classification, profiling, differentiation, authentication or characterization have been a vital and important component of the various studies revealed that the focus has been on plant oils [3–41] by using mainly different types of chromatography, infrared absorption, nuclear magnetic resonance and mass spectrometry techniques. Studies on fish and marine mammal oils are scarce [41–47]. In addition, the literature overview revealed that multivariate techniques, especially principal component analysis (PCA), were the preferred data analysis approaches used in the majority of these oil discrimination studies [3, 5, 8, 10–14, 16, 17, 19–22, 25–28, 30, 34–38, 40, 42–44, 46, 47] and that the potential of simple techniques such as gas chromatography (GC) has not been explored yet in the discrimination of fish and marine mammal oils by using the conventional fatty acid methyl ester (FAME) profiles.
The previous observations seem to indicate that the issue about the importance of techniques for detecting adulteration in n-3 rich oils derived from animals has been overlooked by practitioners during the course of a century and it is a topic of contemporary relevance that needs attention.
The main goal of this study is to evaluate the capability of GC to discriminate between n-3 rich oils derived from fish and marine mammals of different species, brands and grades by using the FAME profiles and PCA. The GC FAME profiles from plant oils such as rapeseed, linseed and soy oils and seven different brands of n-3 supplements are also used in the discrimination process. The discrimination between and within animal oils is studied by using three different data analysis strategies: firstly, the analysis of the full FAME profiles from plant, supplement and animal oils; secondly, the analysis of selected FAME profiles from plant, supplement and animal oils with levels higher than 0.5% of the total composition; thirdly, the analysis of the full FAME profiles from animal oils. It must be mentioned that discrimination studies of n-3 rich oils derived from fish and marine mammals have not been previously reported.
Experimental
Reagents and Samples
Sodium hydroxide, hexane, methanol, boron trifluoride in methanol (20% w/v) and chloroform were purchased from Merck (Darmstadt, Germany). Butylated hydroxytoluene (BHT) and boron trichloride in methanol (14%) were purchased from Sigma–Aldrich Co., USA. FAME pure standards and also model mixture standards 2A and 2B (18:0, 18:1n-9, 18:2n-6, 18:3n-3, 20:4n-6), 3A (18:2n-6, 18:3n-3, 20:4n-6, 22:6n-3), 4A (6:0, 8:0, 10:0, 12:0, 14:0) 6A (16:0, 18:0, 20:0, 22:0, 24:0), 7A (16:1n-7, 16:1n-9, 20:1n-9, 22:1n-11, 24:1n-9) and 14A (13:0, 15:0, 17:0, 19:0, 21:0) were purchased from Nu-Chek Prep (Elysian, MN, USA). Nonadecanoic acid methyl ester (19:0) internal standard was from Fluka (Buchs, Switzerland). De-ionized water was purified in a Milli-Q system (Milli-Q system Millipore, Milford, MA, USA). The fish oils were cod liver oil from Peter Möller, Lysaker, Norway and salmon oil from Havnegater, Sortland, Norway. The two brands of harp seal oils (Pagophilus groenlandicus) were from Rieber Skinn A/S, Bergen, Norway (two refined samples from different batches, designated as RSA1 and RSA2, and one crude sample designated as CSA were provided) and from JFM Sunile A/S, Os, Norway (one refined sample designated as RSB was provided). Whale oil (Balaenoptera acutorostrata) conventionally (WC) and molecularly (WM) distilled were from Myklebust Trading AS, Myklebost, Norway. The plant oils analyzed were soy oil (Mills DA, Sofienberg, Norway), linseed and rapeseed oils (Kinsarvik Naturkost, Bergen, Norway). The seven commercial n-3 supplements obtained from a local pharmacy were Fri Flyt (Vesterålens Naturprodukter AS, Sortland, Norway), Natur-Omega (Naturhuset AS, Vøyenenga, Norway), Møllers dobbel (Peter Möller, Lysaker, Norway), Pikasol (Axellus A/S, Oslo, Norway), Omega-3 Forte (Vitamed, Sarpsborg, Norway), Omega-3 høykonsentrert (Sunkost, Oslo, Norway), El Dorado (Probio Nutraceuticals, Tromsø, Norway). The supplements were designated as K1, K2, K3, K4, K5, K6 and K7, respectively.
FAME Preparation
The FAME preparation protocol has been published elsewhere [48]. Briefly, 50 mg of the sample are mixed with 2 ml BF3/CH3OH and 5 mg of the 19:0 internal standard. The mixture is heated at 100 °C for 1 h and cooled down to room temperature. Aliquots of 1 ml of hexane and 2 ml of H2O are added, vortex-mixed for 15 s, placed in a centrifuge at 3,000 rpm for 2 min and the FAME are then extracted from the upper hexane phase. Depending on the fat content the sample is either concentrated under nitrogen or diluted with hexane and subsequently subjected to GC analysis.
The FAME for every kind of oil (fish, marine mammal and plant) were prepared in triplicate and for the n-3 supplements in duplicate.
Gas Chromatography Instrumentation
Analysis of the FAME was performed on a Perkin-Elmer AutoSystem XL gas chromatograph (Perkin-Elmer, Norwalk, Connecticut) equipped with a liquid autosampler and a flame ionization detector. The FAME samples were analyzed on a CP-Sil 88 capillary column (50 m × 0.32 mm ID 0.2 μm film thickness, Varian, Courtaboeuf, France). Data collection was performed by the Perkin-Elmer TotalChrom Data System software version 6.3. The temperature program was as follows: the oven temperature was held at 60 °C for 1 min, ramped to 160 °C at 25 °C/min, held at 160 °C for 28 min, ramped to 190 °C at 25 °C/min, held at 190 °C for 17 min, ramped to 220 °C at 25 °C/min and finally held at 220 °C for 10 min. Direct on-column injection was used. The injector port temperature was ramped instantaneously from 50 to 250 °C and the detector temperature was 250 °C. The carrier gas was ultra-pure helium at a pressure of 82 KPA. The analysis time was 60 min. This time interval was sufficient to detect FAME with chains from 10 to 24 carbons in length. The FAME peaks were identified by comparison of their retention times with the retention times of highly purified FAME standards. Since the concentration of the internal standard (19:0) is known and its recovery in the different oils was constant, the comparison of the various FAME peak areas with 19:0 peak area is used to calculate the concentration of the FAME in the various oils and supplements.
Principal Component Analysis
Principal component analysis (PCA) is a well documented multivariate method for reducing the dimensionality of a data set by rotating and constructing orthogonal linear combinations of the original variables and projecting the maximum variability onto a new axis also known as principal components (PCs). The results are classified according to the level of information produced by the various combinations of the original variables in a way that the first component (PC1) is the major axis of the points in the p-dimensional space that accounts for the largest variation in the data and consequently it contains the most possible information. The second component (PC2) is perpendicular to PC1 and it defines the next largest amount of variation.
The results as presented in the various tables (Tables 1, 2, 3, 4 and 5) were combined (according to the various approaches to be discussed) and arranged in m × n data matrices where m represents every prepared oil sample and n represents every analyzed fatty acid. The matrices submitted to PCA are standardized by subtracting their means and dividing by their standard deviations in order to construct linear combinations of the predictor variables n that contains the greatest variance. The PCA score and loading plots of the FAME profiles from the various oils were computed with the software package Statgraphics Plus 5.1 (Statistical Graphics Corp.).
Table 1.
FAME concentrations (mg/g) for different brands and grades of seal oils
| Sample | Seal oil | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Manufacturer | A | B | ||||||||||
| Quality | Crude | Refined | Refined | |||||||||
| Designation | CSA | RSA1 (batch 1) | RSA2 (batch 2) | RSB | ||||||||
| Replicate | i | ii | iii | i | ii | iii | i | ii | iii | i | ii | iii |
| 14:0 | 42.8 | 44.3 | 41.5 | 42.2 | 42.5 | 41.6 | 45.4 | 45.9 | 45.9 | 40.2 | 40.3 | 40.1 |
| 15:0 | 2.5 | 2.7 | 2.5 | 2.6 | 2.7 | 2.4 | 2.8 | 2.9 | 2.8 | 2.8 | 2.9 | 2.9 |
| 16:0 | 72.8 | 75.5 | 70.4 | 69.6 | 70.4 | 68.5 | 66.2 | 66.7 | 65.9 | 75.5 | 75.8 | 75.3 |
| 17:0 | 0.5 | 0.5 | 0.5 | 0.4 | 0.4 | 0.4 | 0.7 | 0.7 | 0.7 | 1.7 | 1.7 | 1.7 |
| 18:0 | 10.5 | 10.8 | 10.1 | 10.0 | 10.0 | 9.7 | 8.4 | 8.8 | 8.5 | 12.2 | 12.6 | 11.9 |
| 20:0 | 0.3 | 0.3 | 0.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Total SFA | 129.4 | 134.1 | 125.3 | 124.8 | 126.0 | 122.6 | 123.5 | 125.0 | 123.8 | 132.4 | 133.3 | 131.9 |
| 14:1n-9 | 6.3 | 6.7 | 6.2 | 6.9 | 7.1 | 6.6 | 7.1 | 7.3 | 7.0 | 6.0 | 6.0 | 6.0 |
| 16:1n-9 | 3.5 | 3.8 | 3.5 | 4.3 | 4.3 | 4.0 | 3.8 | 3.9 | 3.9 | 4.6 | 4.8 | 4.5 |
| 16:1n-7 | 143.5 | 148.6 | 138.5 | 149.9 | 151.0 | 148.7 | 155.9 | 157.1 | 155.7 | 147.8 | 148.3 | 147.5 |
| 18:1n-11 | 30.2 | 32.0 | 29.6 | 39.9 | 41.3 | 38.3 | 39.7 | 40.5 | 39.1 | 44.8 | 45.1 | 44.7 |
| 18:1n-9 | 158.0 | 163.5 | 152.3 | 153.1 | 156.2 | 149.8 | 149.6 | 150.1 | 149.1 | 152.6 | 153.3 | 152.0 |
| 18:1n-7 | 38.3 | 38.4 | 38.2 | 38.5 | 38.6 | 38.2 | 35.5 | 35.9 | 35.2 | 42.4 | 42.7 | 42.5 |
| 20:1n-11 | 17.2 | 17.8 | 16.4 | 21.0 | 21.8 | 20.2 | 18.9 | 19.4 | 18.8 | 25.8 | 26.0 | 25.8 |
| 20:1n-9 | 85.2 | 89.1 | 81.1 | 85.5 | 86.4 | 84.4 | 70.5 | 72.3 | 69.8 | 99.6 | 100.3 | 99.9 |
| 20:1n-7 | 5.4 | 5.5 | 5.0 | 5.2 | 5.3 | 5.1 | 4.2 | 4.4 | 4.2 | 6.2 | 6.5 | 6.0 |
| 22:1n-11 | 18.9 | 19.5 | 18.2 | 19.7 | 20.5 | 18.8 | 17.2 | 17.6 | 17.1 | 23.5 | 23.6 | 23.2 |
| 22:1n-9 | 4.7 | 4.7 | 4.2 | 4.7 | 4.8 | 4.4 | 4.0 | 4.1 | 4.0 | 6.1 | 6.2 | 5.9 |
| 24:1n-9 | 4.7 | 4.8 | 4.4 | 4.8 | 4.9 | 4.6 | 5.0 | 5.1 | 5.0 | 4.3 | 4.3 | 4.3 |
| Total MUFA | 515.9 | 534.4 | 497.6 | 533.5 | 542.2 | 523.1 | 511.4 | 517.7 | 508.9 | 563.7 | 567.1 | 562.3 |
| 16:2n-4 | 6.5 | 6.8 | 6.4 | 5.7 | 5.8 | 5.4 | 5.7 | 5.7 | 5.7 | 4.6 | 4.7 | 4.6 |
| 16:3n-3 | 2.4 | 2.6 | 2.4 | 2.0 | 2.1 | 2.0 | 2.0 | 2.1 | 2.0 | 1.6 | 1.6 | 1.6 |
| 16:4n-3 | 4.3 | 4.5 | 4.2 | 3.0 | 3.0 | 2.8 | 3.8 | 4.0 | 3.8 | 3.4 | 3.6 | 3.4 |
| 18:2n-6 | 17.5 | 17.8 | 16.4 | 16.5 | 16.7 | 16.6 | 18.0 | 18.2 | 17.9 | 15.4 | 15.8 | 15.1 |
| 18:3n-3 | 5.0 | 5.1 | 4.7 | 4.6 | 4.7 | 4.5 | 5.0 | 5.0 | 5.0 | 4.5 | 4.7 | 4.4 |
| 18:4n-3 | 13.0 | 13.3 | 12.4 | 11.6 | 12.0 | 10.9 | 12.4 | 13.0 | 12.9 | 9.3 | 9.4 | 9.3 |
| 20:2n-6 | 1.7 | 1.8 | 1.6 | 1.7 | 1.8 | 1.7 | 1.6 | 1.6 | 1.6 | 2.1 | 2.1 | 2.0 |
| 20:4n-6 | 4.9 | 5.1 | 4.7 | 4.2 | 4.5 | 4.2 | 4.9 | 5.0 | 4.8 | 3.6 | 3.7 | 3.5 |
| 20:4n-3 | 4.5 | 4.6 | 4.2 | 4.3 | 4.3 | 4.0 | 4.6 | 4.7 | 4.6 | 3.8 | 3.9 | 3.8 |
| 20:5n-3 | 67.2 | 70.7 | 65.7 | 61.3 | 64.5 | 58.2 | 64.5 | 64.9 | 64.3 | 53.7 | 54.8 | 53.5 |
| 22:5n-3 | 38.1 | 40.3 | 37.2 | 38.6 | 40.0 | 37.0 | 38.1 | 38.5 | 37.9 | 36.6 | 36.8 | 36.5 |
| 22:6n-3 | 82.7 | 86.4 | 78.6 | 76.8 | 81.5 | 74.1 | 88.2 | 90.6 | 86.9 | 57.5 | 58.3 | 57.6 |
| Total n-3 | 217.2 | 227.5 | 209.4 | 202.2 | 212.1 | 193.5 | 218.6 | 222.8 | 217.4 | 170.4 | 173.1 | 170.1 |
| Total n-6 | 24.1 | 24.7 | 22.7 | 22.4 | 23.0 | 22.5 | 24.5 | 24.8 | 24.3 | 21.1 | 21.6 | 20.6 |
| Total PUFA | 247.8 | 259.0 | 238.5 | 230.3 | 240.9 | 221.4 | 248.8 | 253.3 | 247.4 | 196.1 | 199.4 | 195.3 |
SFA saturated fatty acids, MUFA monounsaturated fatty acids, PUFA polyunsaturated fatty acids
Table 2.
FAME concentrations (mg/g) for whale (different grades) and fish (different species) oils
| Sample | Whale oil | Fish oil | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Quality | Conventionally distilled | Molecularly distilled | Cod liver | Salmon | ||||||||
| Designation | WC | WM | CL | SA | ||||||||
| Replicate | i | ii | iii | i | ii | iii | i | ii | iii | i | ii | iii |
| 14:0 | 50.5 | 50.6 | 50.4 | 47.8 | 48.1 | 47.5 | 33.7 | 33.6 | 33.6 | 41.1 | 41.8 | 40.5 |
| 15:0 | 3.4 | 3.5 | 3.3 | 3.6 | 3.7 | 3.3 | 3.1 | 3.2 | 3.0 | 3.9 | 3.9 | 3.8 |
| 16:0 | 74.5 | 75.2 | 72.7 | 95.3 | 95.8 | 94.6 | 91.9 | 92.6 | 91.0 | 130.5 | 133.4 | 127.5 |
| 17:0 | 4.9 | 5.0 | 4.8 | 3.7 | 4.0 | 3.7 | 5.8 | 5.8 | 5.7 | 2.7 | 2.8 | 2.5 |
| 18:0 | 16.4 | 16.1 | 15.6 | 23.5 | 23.9 | 23.4 | 18.5 | 19.1 | 17.8 | 30.5 | 31.4 | 29.8 |
| 20:0 | 0.5 | 0.5 | 0.5 | 0.7 | 0.7 | 0.6 | 0.0 | 0.0 | 0.0 | 1.8 | 1.9 | 1.7 |
| 22:0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.7 | 1.8 | 1.7 |
| 24:0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.7 | 1.8 | 1.6 | 0.0 | 0.0 | 0.0 |
| Total SFA | 150.2 | 150.9 | 147.3 | 174.6 | 176.2 | 173.1 | 154.7 | 156.1 | 152.7 | 212.2 | 217.0 | 207.5 |
| 14:1n-9 | 3.9 | 3.8 | 3.9 | 3.5 | 3.7 | 3.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16:1n-9 | 3.2 | 3.2 | 3.1 | 3.5 | 3.6 | 3.5 | 4.3 | 4.6 | 4.3 | 2.8 | 2.9 | 2.6 |
| 16:1n-7 | 74.0 | 75.2 | 71.8 | 65.1 | 65.7 | 64.6 | 60.7 | 62.8 | 58.6 | 39.6 | 39.9 | 39.1 |
| 18:1n-11 | 18.2 | 18.0 | 17.4 | 19.7 | 19.8 | 19.4 | 13.6 | 13.7 | 13.6 | 3.9 | 4.0 | 3.8 |
| 18:1n-9 | 147.0 | 147.3 | 146.1 | 136.6 | 137.1 | 135.9 | 122.7 | 123.8 | 121.4 | 242.1 | 243.2 | 240.9 |
| 18:1n-7 | 21.4 | 21.3 | 20.9 | 25.5 | 25.6 | 25.3 | 34.9 | 35.1 | 34.5 | 29.5 | 30.2 | 28.6 |
| 20:1n-11 | 20.9 | 21.6 | 20.2 | 21.7 | 21.8 | 21.4 | 10.2 | 10.3 | 10.0 | 4.6 | 4.8 | 4.6 |
| 20:1n-9 | 96.9 | 98.1 | 94.8 | 128.4 | 129.1 | 127.6 | 77.9 | 78.2 | 77.7 | 43.1 | 43.5 | 42.7 |
| 20:1n-7 | 2.1 | 2.1 | 2.0 | 2.7 | 2.8 | 2.7 | 3.2 | 3.3 | 3.2 | 2.5 | 2.7 | 2.4 |
| 22:1n-11 | 94.4 | 96.6 | 91.3 | 120.8 | 122.1 | 119.4 | 50.0 | 50.0 | 49.9 | 39.2 | 39.9 | 38.8 |
| 22:1n-9 | 7.0 | 7.1 | 6.7 | 10.5 | 10.9 | 10.1 | 6.0 | 6.1 | 5.9 | 7.5 | 7.9 | 7.3 |
| 24:1n-9 | 6.9 | 7.1 | 6.8 | 6.5 | 7.0 | 6.4 | 7.9 | 7.9 | 7.9 | 8.1 | 8.4 | 7.8 |
| Total MUFA | 495.9 | 501.4 | 485.0 | 544.5 | 549.2 | 539.7 | 391.4 | 395.8 | 387.0 | 422.9 | 427.4 | 418.6 |
| 16:2n-4 | 3.6 | 3.6 | 3.5 | 2.8 | 3.0 | 2.8 | 4.2 | 4.3 | 4.0 | 3.7 | 3.9 | 3.7 |
| 16:3n-3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.4 | 3.4 | 3.4 | 1.9 | 2.0 | 1.8 |
| 16:4n-3 | 2.6 | 2.6 | 2.5 | 0.0 | 0.0 | 0.0 | 4.6 | 4.7 | 4.3 | 3.2 | 3.3 | 3.0 |
| 18:2n-6 | 19.4 | 19.7 | 19.2 | 17.4 | 17.6 | 17.0 | 18.5 | 18.5 | 18.4 | 81.4 | 82.3 | 80.4 |
| 18:3n-3 | 12.6 | 12.7 | 12.4 | 10.9 | 11.1 | 10.7 | 7.8 | 7.8 | 7.5 | 30.4 | 31.0 | 29.9 |
| 18:4n-3 | 27.6 | 27.9 | 26.3 | 19.4 | 19.8 | 18.7 | 24.5 | 25.1 | 23.7 | 9.2 | 9.3 | 8.8 |
| 20:3n-3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.3 | 3.4 | 3.2 |
| 20:2n-6 | 3.0 | 3.1 | 2.8 | 3.5 | 3.6 | 3.4 | 2.6 | 2.6 | 2.5 | 6.9 | 7.1 | 6.6 |
| 20:3n-6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.6 | 1.7 | 1.5 |
| 20:4n-6 | 3.8 | 3.9 | 3.6 | 2.8 | 3.0 | 2.8 | 6.5 | 6.5 | 6.5 | 3.4 | 3.6 | 3.4 |
| 20:4n-3 | 14.1 | 14.1 | 14.0 | 11.1 | 11.3 | 10.7 | 8.0 | 8.0 | 7.9 | 10.7 | 11.4 | 10.3 |
| 20:5n-3 | 46.5 | 47.2 | 45.3 | 35.1 | 36.1 | 33.9 | 106.7 | 108.1 | 105.1 | 39.1 | 40.7 | 37.7 |
| 22:5n-3 | 23.0 | 23.0 | 22.9 | 20.5 | 21.1 | 20.3 | 16.4 | 16.6 | 16.0 | 21.9 | 22.4 | 21.3 |
| 22:6n-3 | 76.7 | 76.3 | 75.1 | 48.3 | 50.7 | 47.3 | 145.6 | 149.6 | 141.2 | 52.3 | 54.0 | 50.4 |
| Total n-3 | 203.1 | 203.8 | 198.5 | 145.3 | 150.1 | 141.6 | 317.0 | 323.3 | 309.1 | 172.0 | 177.5 | 166.4 |
| Total n-6 | 26.2 | 26.7 | 25.6 | 23.7 | 24.2 | 23.2 | 27.6 | 27.6 | 27.4 | 93.3 | 94.7 | 91.9 |
| Total PUFA | 232.9 | 234.1 | 227.6 | 171.8 | 177.3 | 167.6 | 348.8 | 355.2 | 340.5 | 269.0 | 276.1 | 262.0 |
SFA saturated fatty acids, MUFA monounsaturated fatty acids, PUFA polyunsaturated fatty acids
Table 3.
FAME concentrations (mg/g) for different brands of n-3 supplements
| Sample | Supplements | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Designation | K1 | K2 | K3 | K4 | K5 | K6 | K7 | |||||||
| Replicate | i | ii | i | ii | i | ii | i | ii | i | ii | i | ii | i | ii |
| 14:0 | 2.6 | 2.5 | 0.0 | 0.0 | 22.2 | 22.2 | 1.3 | 1.2 | 3.0 | 3.1 | 2.5 | 2.6 | 2.7 | 1.2 |
| 15:0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.4 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16:0 | 22.2 | 22.4 | 44.7 | 44.4 | 58.5 | 58.8 | 24.2 | 23.9 | 22.7 | 22.7 | 12.4 | 12.6 | 22.7 | 15.3 |
| 17:0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.7 | 2.1 |
| 18:0 | 29.7 | 29.9 | 22.8 | 22.7 | 18.8 | 19.2 | 31.6 | 31.5 | 27.0 | 26.5 | 14.2 | 14.6 | 29.8 | 22.1 |
| 20:0 | 3.3 | 3.4 | 1.9 | 2.0 | 2.5 | 2.3 | 4.8 | 4.8 | 9.3 | 9.2 | 4.3 | 4.3 | 3.4 | 6.6 |
| 22:0 | 1.0 | 0.9 | 0.0 | 0.0 | 0.9 | 0.8 | 0.0 | 0.0 | 2.4 | 2.4 | 0.0 | 0.0 | 0.0 | 1.2 |
| 24:0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.1 | 1.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Total SFA | 58.8 | 59.1 | 69.4 | 69.1 | 105.4 | 106.0 | 61.9 | 61.4 | 64.4 | 63.9 | 33.4 | 34.1 | 58.8 | 59.1 |
| 16:1n-9 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16:1n-7 | 8.7 | 8.5 | 0.0 | 0.0 | 26.3 | 25.9 | 8.6 | 8.3 | 8.4 | 8.4 | 7.2 | 7.4 | 8.7 | 7.9 |
| 18:1n-9 | 62.2 | 62.6 | 175.0 | 174.1 | 46.3 | 45.4 | 60.1 | 59.3 | 51.9 | 51.0 | 39.5 | 40.0 | 62.7 | 50.7 |
| 18:1n-7 | 18.3 | 18.4 | 8.5 | 8.6 | 14.5 | 13.9 | 21.9 | 21.5 | 20.2 | 19.9 | 13.7 | 13.7 | 18.7 | 16.5 |
| 20:1n-11 | 1.8 | 1.7 | 2.9 | 2.8 | 2.0 | 1.9 | 2.5 | 2.4 | 1.2 | 1.1 | 1.4 | 1.3 | 1.8 | 1.1 |
| 20:1n-9 | 13.5 | 13.5 | 0.0 | 0.0 | 13.9 | 13.7 | 24.2 | 23.8 | 23.2 | 23.0 | 19.2 | 19.5 | 13.9 | 23.7 |
| 20:1n-7 | 4.5 | 4.5 | 0.0 | 0.0 | 3.0 | 3.1 | 4.3 | 4.3 | 3.2 | 3.1 | 3.4 | 3.5 | 4.5 | 2.8 |
| 22:1n-11 | 11.2 | 11.6 | 0.0 | 0.0 | 20.6 | 19.4 | 15.5 | 15.3 | 8.9 | 8.5 | 10.8 | 10.7 | 11.5 | 8.2 |
| 22:1n-9 | 1.5 | 1.5 | 0.0 | 0.0 | 2.8 | 2.6 | 3.3 | 3.2 | 5.2 | 5.1 | 4.7 | 4.6 | 1.5 | 5.5 |
| 24:1n-9 | 9.3 | 9.4 | 0.0 | 0.0 | 5.0 | 5.6 | 11.4 | 11.3 | 12.2 | 12.1 | 16.3 | 17.2 | 11.9 | 14.5 |
| Total MUFA | 131.0 | 131.7 | 186.4 | 185.5 | 135.6 | 132.6 | 151.8 | 149.4 | 134.4 | 132.2 | 116.2 | 117.9 | 131.0 | 131.7 |
| 16:2n-4 | 1.6 | 1.6 | 0.0 | 0.0 | 4.4 | 4.8 | 1.0 | 1.1 | 1.1 | 1.1 | 1.1 | 1.2 | 1.9 | 1.0 |
| 16:3n-3 | 2.4 | 2.3 | 0.0 | 0.0 | 6.2 | 6.0 | 1.5 | 1.6 | 1.7 | 1.6 | 1.5 | 1.4 | 2.0 | 1.2 |
| 18:2n-6 | 6.9 | 7.0 | 259.9 | 258.8 | 5.7 | 5.3 | 8.2 | 8.2 | 7.0 | 6.7 | 5.0 | 5.4 | 7.2 | 7.2 |
| 18:3n-3 | 3.7 | 3.7 | 312.5 | 312.5 | 3.5 | 3.3 | 5.8 | 5.7 | 4.5 | 4.5 | 3.3 | 3.1 | 3.7 | 3.9 |
| 18:4n-3 | 16.4 | 16.5 | 0.0 | 0.0 | 13.7 | 13.6 | 21.1 | 21.0 | 20.6 | 20.3 | 13.1 | 12.8 | 16.4 | 16.7 |
| 20:2n-6 | 2.1 | 2.1 | 0.0 | 0.0 | 1.6 | 1.6 | 2.5 | 2.6 | 2.1 | 2.2 | 2.1 | 2.2 | 2.2 | 2.0 |
| 20:3n-6 | 1.8 | 2.0 | 0.0 | 0.0 | 1.4 | 1.5 | 2.3 | 2.3 | 2.5 | 2.5 | 2.4 | 2.6 | 1.8 | 2.2 |
| 20:4n-6 | 12.9 | 13.1 | 0.0 | 0.0 | 9.4 | 8.9 | 17.7 | 17.6 | 15.7 | 15.4 | 15.3 | 14.9 | 12.9 | 14.0 |
| 20:4n-3 | 10.5 | 10.5 | 0.0 | 0.0 | 7.7 | 8.0 | 14.7 | 14.5 | 16.2 | 16.0 | 14.7 | 14.5 | 10.5 | 15.1 |
| 20:5n-3 | 242.6 | 245.0 | 0.0 | 0.0 | 174.9 | 174.2 | 283.8 | 281.5 | 276.4 | 273.1 | 260.3 | 250.1 | 239.8 | 238.1 |
| 22:5n-3 | 24.9 | 25.2 | 0.0 | 0.0 | 25.6 | 26.9 | 28.9 | 28.7 | 35.8 | 35.2 | 42.1 | 40.8 | 24.7 | 36.6 |
| 22:6n-3 | 168.0 | 169.4 | 0.0 | 0.0 | 167.8 | 182.4 | 175.1 | 173.6 | 175.3 | 173.0 | 198.6 | 182.9 | 162.8 | 172.4 |
| Total n-3 | 468.5 | 472.6 | 312.5 | 312.5 | 399.4 | 414.4 | 530.9 | 526.6 | 530.5 | 523.7 | 533.6 | 505.6 | 459.9 | 484.0 |
| Total n-6 | 23.7 | 24.2 | 259.9 | 258.8 | 18.1 | 17.3 | 30.7 | 30.7 | 27.3 | 26.8 | 24.8 | 25.1 | 24.1 | 25.4 |
| Total PUFA | 493.8 | 498.4 | 572.4 | 571.3 | 421.9 | 436.5 | 562.6 | 558.4 | 558.9 | 551.6 | 559.5 | 531.9 | 493.8 | 498.4 |
SFA saturated fatty acids, MUFA monounsaturated fatty acids, PUFA polyunsaturated fatty acids
Table 4.
FAME concentrations (mg/g) for different plant oils
| Sample | Plant oil | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Name | Soy | Rapeseed | Linseed | ||||||
| Designation | SO | RP | LN | ||||||
| Replicate | i | ii | iii | i | ii | iii | i | ii | iii |
| 16:0 | 89.5 | 90.3 | 89.0 | 35.8 | 36.2 | 34.5 | 40.5 | 41.3 | 39.7 |
| 18:0 | 28.2 | 28.5 | 27.9 | 15.0 | 15.4 | 14.5 | 33.9 | 34.9 | 32.9 |
| 20:0 | 4.5 | 4.6 | 4.5 | 5.4 | 5.5 | 5.2 | 3.1 | 3.1 | 3.0 |
| 22:0 | 4.4 | 4.4 | 4.4 | 2.7 | 2.8 | 2.7 | 0.9 | 0.9 | 0.9 |
| 24:0 | 1.5 | 1.6 | 1.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Total SFA | 128.1 | 129.4 | 127.3 | 58.9 | 59.9 | 56.9 | 78.4 | 80.2 | 76.5 |
| 16:1n-7 | 0.0 | 0.0 | 0.0 | 1.7 | 1.7 | 1.6 | 0.0 | 0.0 | 0.0 |
| 18:1n-9 | 225.4 | 225.6 | 225.4 | 565.3 | 567.5 | 563.0 | 194.6 | 195.8 | 193.3 |
| 18:1n-7 | 13.6 | 13.7 | 13.4 | 24.7 | 25.2 | 24.1 | 5.5 | 5.5 | 5.4 |
| 20:1n-9 | 2.1 | 2.1 | 2.0 | 10.1 | 10.4 | 9.7 | 0.0 | 0.0 | 0.0 |
| Total MUFA | 241.1 | 241.4 | 240.8 | 601.8 | 604.8 | 598.4 | 200.1 | 201.3 | 198.7 |
| 18:2n-6 | 494.0 | 496.1 | 494.0 | 172.7 | 176.8 | 168.4 | 133.4 | 134.3 | 132.5 |
| 18:3n-3 | 51.9 | 52.0 | 51.9 | 84.6 | 85.5 | 83.6 | 506.1 | 507.3 | 504.9 |
| Total n-3 | 51.9 | 52.0 | 51.9 | 84.6 | 85.5 | 83.6 | 506.1 | 507.3 | 504.9 |
| Total n-6 | 494.0 | 496.1 | 494.0 | 172.7 | 176.8 | 168.4 | 133.4 | 134.3 | 132.5 |
| Total PUFA | 545.9 | 548.1 | 545.9 | 257.3 | 262.3 | 252.0 | 639.5 | 641.6 | 637.4 |
SFA saturated fatty acids, MUFA monounsaturated fatty acids, PUFA polyunsaturated fatty acids
Table 5.
FAME concentrations (mg/g) for conventionally distilled whale oil (WC) adulterated with of cod liver oil (CL)
| Sample | Mixtures of conventional distilled whale oil (WC) and cod liver oil (CL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| WC:CL (%) | 90:10 | 80:20 | 50:50 | ||||||
| Replicate | i | ii | iii | i | ii | iii | i | ii | iii |
| 14:0 | 48.0 | 48.1 | 48.1 | 47.5 | 47.4 | 47.9 | 42.5 | 42.3 | 41.9 |
| 15:0 | 3.4 | 3.4 | 3.3 | 3.3 | 3.4 | 3.3 | 3.3 | 3.3 | 3.2 |
| 16:0 | 77.0 | 77.0 | 76.1 | 78.0 | 78.1 | 77.9 | 83.2 | 83.4 | 83.2 |
| 17:0 | 5.1 | 5.1 | 5.1 | 5.1 | 5.2 | 5.1 | 5.4 | 5.4 | 5.4 |
| 18:0 | 16.3 | 16.4 | 16.2 | 16.5 | 16.6 | 16.5 | 17.2 | 17.4 | 17.2 |
| 20:0 | 0.5 | 0.5 | 0.5 | 0.4 | 0.4 | 0.4 | 0.3 | 0.3 | 0.3 |
| 24:0 | 0.2 | 0.2 | 0.2 | 0.4 | 0.4 | 0.4 | 0.9 | 0.9 | 0.9 |
| Total SFA | 150.5 | 150.7 | 149.5 | 151.2 | 151.5 | 151.5 | 152.8 | 153.0 | 152.1 |
| 14:1n-9 | 3.3 | 3.3 | 3.3 | 3.1 | 3.1 | 3.0 | 1.9 | 1.9 | 1.9 |
| 16:1n-9 | 3.3 | 3.4 | 3.3 | 3.5 | 3.5 | 3.5 | 3.8 | 3.8 | 3.8 |
| 16:1n-7 | 72.1 | 72.1 | 71.8 | 71.6 | 71.8 | 71.1 | 67.1 | 67.9 | 68.2 |
| 18:1n-11 | 17.6 | 17.6 | 17.5 | 17.1 | 17.2 | 17.1 | 15.8 | 15.9 | 15.9 |
| 18:1n-9 | 144.5 | 145.9 | 142.8 | 142.7 | 143.5 | 142.2 | 135.0 | 136.3 | 136.3 |
| 18:1n-7 | 22.5 | 22.7 | 22.5 | 23.9 | 24.1 | 23.8 | 27.9 | 28.1 | 27.9 |
| 20:1n-11 | 19.9 | 19.9 | 19.9 | 18.8 | 18.8 | 18.8 | 15.6 | 15.6 | 15.6 |
| 20:1n-9 | 95.2 | 95.4 | 95.2 | 93.4 | 93.5 | 93.3 | 87.7 | 87.9 | 87.7 |
| 20:1n-7 | 2.2 | 2.2 | 2.2 | 2.3 | 2.3 | 2.3 | 2.7 | 2.7 | 2.7 |
| 22:1n-11 | 89.7 | 90.1 | 89.5 | 85.3 | 85.7 | 85.1 | 71.8 | 72.4 | 71.8 |
| 22:1n-9 | 6.9 | 6.9 | 6.8 | 6.8 | 6.8 | 6.7 | 6.5 | 6.5 | 6.5 |
| 24:1n-9 | 7.0 | 7.1 | 7.0 | 7.1 | 7.2 | 7.1 | 7.4 | 7.5 | 7.4 |
| Total MUFA | 484.2 | 486.6 | 481.8 | 475.6 | 477.5 | 474.0 | 443.2 | 446.5 | 445.7 |
| 16:2n-4 | 3.6 | 3.6 | 3.6 | 3.7 | 3.7 | 3.7 | 3.9 | 3.9 | 3.9 |
| 16:3n-3 | 0.3 | 0.3 | 0.3 | 0.7 | 0.7 | 0.7 | 1.7 | 1.7 | 1.7 |
| 16:4n-3 | 2.8 | 2.8 | 2.7 | 3.0 | 3.0 | 2.9 | 3.5 | 3.6 | 3.5 |
| 18:2n-6 | 19.4 | 19.4 | 19.4 | 19.3 | 19.3 | 19.3 | 19.0 | 19.0 | 19.0 |
| 18:3n-3 | 12.0 | 12.2 | 12.0 | 11.6 | 11.7 | 11.5 | 10.0 | 10.3 | 10.0 |
| 18:4n-3 | 27.2 | 27.4 | 27.0 | 26.9 | 27.1 | 26.7 | 25.9 | 26.3 | 25.9 |
| 20:2n-6 | 2.9 | 2.9 | 2.9 | 2.9 | 2.9 | 2.9 | 2.8 | 2.8 | 2.8 |
| 20:4n-6 | 4.0 | 4.0 | 4.0 | 4.3 | 4.3 | 4.3 | 5.1 | 5.2 | 5.1 |
| 20:4n-3 | 13.5 | 13.5 | 13.4 | 12.9 | 12.9 | 12.8 | 11.0 | 11.1 | 11.0 |
| 20:5n-3 | 53.2 | 53.5 | 53.1 | 59.3 | 59.5 | 59.1 | 77.2 | 77.6 | 77.2 |
| 22:5n-3 | 22.3 | 22.4 | 22.3 | 21.7 | 21.7 | 21.6 | 19.6 | 19.7 | 19.6 |
| 22:6n-3 | 83.8 | 84.6 | 83.5 | 90.8 | 91.5 | 90.4 | 111.3 | 112.3 | 109.2 |
| Total n-3 | 215.1 | 216.7 | 214.3 | 226.9 | 228.1 | 225.7 | 260.2 | 262.6 | 258.1 |
| Total n-6 | 26.3 | 26.3 | 26.3 | 26.5 | 26.5 | 26.5 | 26.9 | 27.0 | 26.9 |
| Total PUFA | 245.0 | 246.6 | 244.2 | 257.1 | 258.3 | 255.9 | 291.0 | 293.5 | 288.9 |
SFA saturated fatty acids, MUFA monounsaturated fatty acids, PUFA polyunsaturated fatty acids
Results
The fish, marine mammal and plant oils were prepared in triplicate and the n-3 supplements in duplicate. The triplicate and duplicate lipid profiles from the various injected oil samples, expressed as mg FAME/g sample, are presented in Tables 1, 2, 3, 4 and 5. The individual profiles were arranged in a data matrix consisting of 47 rows representing the various analyzed oils with their respective replicates and 34 columns representing the individual FAME detected by GC. The 34 individual FAME profiles were: 14:0, 14:1n-9, 15:0, 16:0, 16:1n-9, 16:1n-7, 17:0, 16:2n-4, 18:0, 16:3n-3, 18:1n-11, 18:1n-9, 18:1n-7, 16:4n-3, 18:2n-6, 20:0, 18:3n-3, 20:1n-11, 20:1n-9, 20:1n-7, 18:4n-3, 20:2n-6, 20:3n-6, 22:0, 20:3n-3, 20:4n-6, 22:1n-11, 22:1n-9, 20:4n-3, 20:5n-3, 24:0, 24:1n-9, 22:5n-3, 22:6n-3.
Discrimination by Using the Full FAME Profiles from Plant, Supplement and Marine Animal Oils
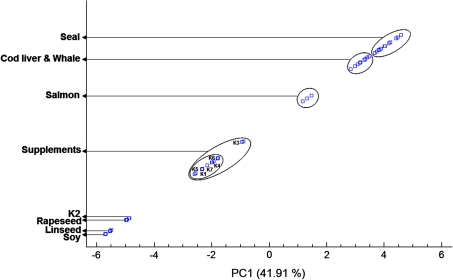
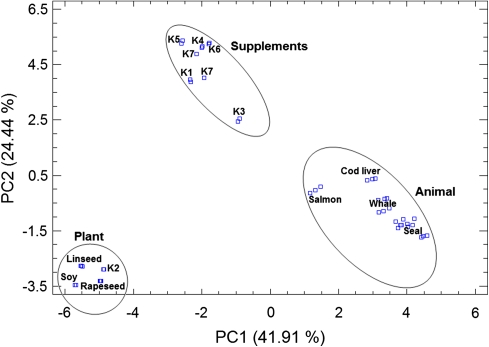
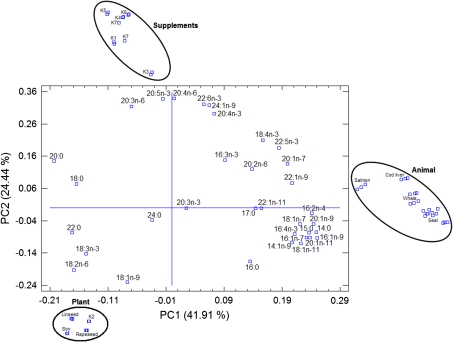
The 47 × 34 matrix was submitted to PCA as a data exploration technique and a total of six components (PCs) were extracted and grouped in decreasing order of variance. The first component (PC1) which explains 41.91% of the total variation can be used to discriminate the oils according to their nature as is shown in Fig. 1. A plot of the scores of the two first components (Fig. 2), which explain 66.35% of the data variation, differentiates basically the same number of groups and sub-groups found in Fig. 1. Besides, the loadings of the two first components, were plotted to investigate the relationship between the various FAME (Fig. 3).
Fig. 1.
PC1 score plot for the different kinds of oils obtained after computing a 47 × 34 (samples × FAME profiles) data matrix. The different supplements and providers are designated by numbered letter Ks. For details regarding the providers see “Experimental”
Fig. 2.
PC1 and PC2 score plot for the different kinds of oils obtained after computing a 47 × 34 (samples × FAME profiles) data matrix. The different supplements and providers are designated by numbered letter Ks. For details regarding the providers see “Experimental”
Fig. 3.
FAME loading plot for PC1 and PC2 and its relationship to the scores portrayed in Fig. 2. The different supplements and providers are designated by numbered letter Ks. For details regarding the providers see “Experimental”
The six PCs computed by using the 47 × 34 matrix were plotted against each other to produce two and three dimensional PC scores graphs and consequently explore the capability of these PCs to discriminate confidently within the marine animal oils. Unfortunately, clear and well-defined patterns that allow one to differentiate the various oils and grades were not observed in any of the graphical representations, hence a data reduction, based on selected FAME profiles, was implemented.
Discrimination by Using Selected FAME Profiles from Plant, Supplement and Marine Animal Oils
FAME data reduction has been used in the discrimination of oils derived from one fish species (cod liver oil) by selecting the 15 FAME with levels higher than 1% of the total composition [42]. Considering that in the present study the 34 FAME or variables are given in mg/g and arranged in columns for PCA purposes (47 × 34), it was decided to discard all the FAME columns with average values <5 mg/g (<0.5% of the total averaged FAME profile), in that way 19 FAME profiles were retained (14:0, 16:0, 16:1n-7, 18:0, 18:1n-11, 18:1n-9, 18:1n-7, 18:2n-6, 18:3n-3, 20:1n-11, 20:1n-9, 18:4n-3, 20:4n-6, 22:1n-11, 20:4n-3, 20:5n-3, 24:1n-9, 22:5n-3, 22:6n-3). A new data matrix of size 47 × 19 was submitted to PCA and three PCs, explaining 85.29% of the total data variability, were computed. The three PCs were used to generate various two and pseudo-three dimensional score plots which essentially showed a clear discrimination between plant, supplements and marine animal oils as already observed in Figs. 1, 2.
Discrimination by Using Full FAME Profiles from Marine Animal Oils
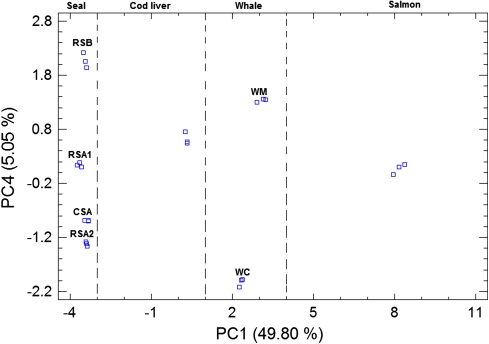
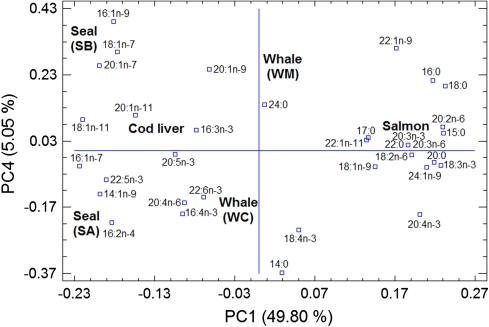
The contribution of the supplement and plant oils was removed as an alternative approach to discriminate the various marine oils. A 24 × 34 matrix was constructed by using the data presented in Tables 1, 2 and submitted to PCA. A considerable percentage of the variability of this matrix (97.58%) was explained by four extracted PCs (49.80, 22.65, 20.08 and 5.05%). The score plot of PC1 and PC4 (Fig. 4) indicates that it is possible to discriminate within the four different types of animal oils by using these two components.
Fig. 4.
PC1 and PC4 score plot for the fish and marine mammal oils obtained after computing a 24 × 34 (samples × FAME profiles) data matrix. RSA1 and RSA2 are refined seal oils from provider A and from different batches. CSA is crude seal oil from provider A. RSB is refined seal oil from provider B. WM and WC are molecularly and conventionally distilled whale oils from the same provider. For details regarding the providers see “Experimental”
Discrimination Between Genuine and Adulterated Oils
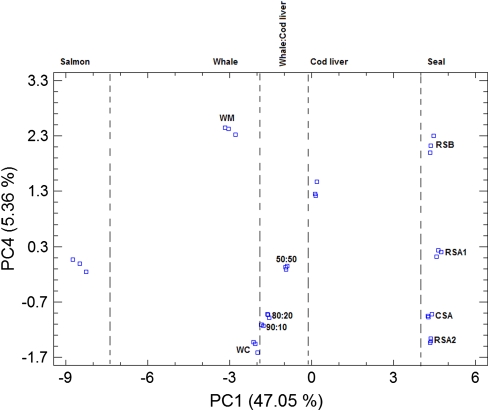
Further assessment of the capability of GC for discriminating between pure and adulterated oils, was achieved using conventionally distilled whale oil (WC) debased by cod liver oil in the proportions 90:10, 80:20 and 50:50. The FAME profiles of the mixtures prepared in triplicate (Table 5) along with the profiles of the marine animal oils described in Tables 1 and 2 were submitted to PCA and four PCs extracted, explaining 47.05, 25.08, 20.19 and 5.36% of the data variability, respectively.
Discussion
Discrimination by Using the Full FAME Profiles from Plant, Supplement and Marine Animal Oils
A graph of PC1 for the various oil samples (Fig. 1) shows that different kinds of oils can be basically differentiated along the PC1 axis. Specifically, the scores of the analyzed plant and animal oils grouped themselves at opposite ends of the PC1 axis while the scores of six supplements (K1 and K3–7) in the middle of this axis. The scores for K2 overlaps those from rapeseed oil, hence it is likely that this supplement contains this particular oil. In addition, the supplements as well as the animals exhibit some sub-groups which discriminate supplement K3 in the former and salmon oil in the latter. These latent sub-structures are attributed to the consistently high levels of 16:0 and 16:1n-7 in supplement K3 (on average three times higher compared to the others supplements of this group) and the high levels of 18:1n-9, 18:2n-6 and 18:3n-3 in the salmon oil (on average 1.7, 4.7 and 5.0 times higher than the rest of marine animal oils, respectively).
The score plot of the first two PC (Fig. 2) which explained 41.91 and 24.44%, respectively, of the total variability of the different types of oil FAME profiles allowed concluding that PC1 discriminates in effect between animal and plant oils while PC2 differentiates between supplements and plant oils. Besides, this score plot revealed that in addition to rapeseed oil, supplement K2 also contains linseed oil due to the proximity of their scores. This proximity was constantly observed when the scores of any of the six PCs were plotted against each other. The presence of rapeseed and linseed oils in the composition of K2 was confirmed by searching the webpage of the manufacturer of this particular supplement. The PC1 versus PC2 plot (Fig. 2) revealed some variability in the individual scores for K7 which could be attributed to experimental errors, indicating the importance of replication in discrimination studies.
The plot of the loadings of the two first components, expressing the relationship between the various FAME (Fig. 3) showed the lack of correlation between 20:5n-3 and 18:1n-9 on the PC2 axis, indicating that none of the plant oils studied in the present investigation are present in the composition of supplements K1, K3, K4, K5, K6 and K7. These observations are in agreement with the various manufacturers who have reported some special developed oils (name not disclosed), refined fish oil (from non-specified origin), gelatin from pork, etc. among the various constituents of their supplements rather than plant oils.
The superimposition of the three main clusters from Fig. 2 on the loading plot (Fig. 3) demonstrates unequivocally that the observed inverse correlation between 20:5n-3 (positive PC2 loading value) and 18:1n-9 (negative PC2 loading value) is responsible for the discrimination between supplements (with scores highly associated to 20:5n-3) and plant oils (with scores highly associated to 18:1n-9). Similarly, the lack of correlation between 16:1n-9, 14:0, 20:1n-9, 16:2n-4 (positive PC1 loading values) and 22:0, 18:2n-6 (negative PC1 loading values) is responsible for discriminating between animal and plant oils.
Discrimination by Using Selected FAME Profiles from Plant, Supplement and Marine Animal Oils
The discrimination patterns of the various two and pseudo-three dimensional score plots obtained after performing PCA on selected FAME profiles were basically similar to those obtained by using the full FAME profiles (Figs. 1, 2). However, the graphs consistently misclassified supplement K2 as containing a mixture of the three plant oils (soy, linseed and rapeseed oil) while in fact only linseed and rapeseed oil are present in this particular supplement. This result indicates that PCA on full FAME profiles outperforms the proposed data reduction approach for supplement classification. The various plots generated with the aforementioned three PCs were also unable to establish a clear distinction between the various species, brands and grades of animal oils; hence a further discrimination study was carried out by using only the full FAME profiles derived from marine oils.
Discrimination by Using Full FAME Profiles from Marine Animal Oils
The PC1 and PC4 score plot (Fig. 4) shows that PC1 can discriminate between the four different types of animal oils, namely seal oil, cod liver oil, whale oil and salmon oil and that PC4 can discriminate effectively within every animal oil species and their alleged qualities. For instance, it is observed that the two different batches of refined seal oils from manufacturer-A (designated as RSA1 and RSA2 in Fig. 4) display positive and negative PC4 score values, respectively, indicating some differences between them. In addition, the crude and the refined seal oils from the same manufacturer and designated as CSA and RSA2 in Fig. 4, exhibit negative scores values, indicating a correlation between these two oils regardless their alleged qualities. The discrimination between the seal oils from different manufacturers namely Rieber Skinn (SA) and JFM Sunile (SB) is observed in Fig. 4. The variables responsible for the discrimination between the manufactures are visualized by means of the PC1 and PC4 loading plot (Fig. 5). The higher contents of 16:1n-9, 18:1n-7, 20:1n-7 and the lower contents of 16:2n-4 in SB compared to SA are the main fatty acids that contribute to distinguish SA from SB. Similarly, Fig. 4 shows the unmistakably differentiation between molecularly distilled whale oil (WM) from conventionally distilled whale oil (WC) which is mainly due to the slightly higher levels of 20:1n-9, the lower levels of 22:6n-3 and 18:4n-3 as well as the lack of 16:4n-3 in WM compared to WC. The clear-cut distinction between cod liver and salmon oil along the PC1 axis (Fig. 4) is mainly attributed to the contribution of several fatty acids such as 18:1n-11 and 16:1n-7 at negative PC1 values and 20:2n-6 and 18:3n-3 at positive PC1 values as indicated in Fig. 5.
Fig. 5.
FAME loading plot for PC1 and PC4 and its relationship to the scores portrayed in Fig. 4
Discrimination Between Genuine and Adulterated Oils
The PC1and PC4 score plot (Fig. 6) demonstrates that it is possible to discriminate adulterated from genuine whale oil samples. The information retained by PC1 is mainly connected with the nature of the various oils. For instance, five specific regions can be visualized along the PC1 axis (salmon, whale, whale + cod liver, cod liver and seal) while the information retained by PC4 is mainly connected with the discrimination within animal oil species, their alleged qualities and the various proportions of cod liver oil used to adulterate genuine whale oil.
Fig. 6.
PC1 and PC4 score plot for the various genuine marine oils and three samples of adulterated whale oil (WC) with cod liver oil at different proportions (90:10, 80:20 and 50:50). The PCs were obtained after computing a 33 × 34 data matrix. RSA1 and RSA2 are refined seal oils from provider A and from different batches. CSA is crude seal oil from provider A. RSB is refined seal oil from provider B. WM and WC are molecularly and conventionally distilled whale oils from the same provider. For details regarding the providers see “Experimental”. PC1 and PC4 score plot for genuine and adulterated fish and marine mammal oils by using the full FAME profiles
In conclusion, the different approaches used in the discrimination process indicated that PCA on the full FAME profiles is the best strategy to discriminate between the various oils considered in this study. Considering that n-3 rich oils derived from animals are highly regarded as alternative medicines worldwide, the potentiality of unexplored single or coupled techniques for authentication and discrimination of these kinds of oils should be investigated to prevent fraudulent practices.
Acknowledgments
The European Commission in the context of the Erasmus Mundus Program and The Norwegian Research Council (SIP project NRF 173534/I30) are gratefully acknowledged for financial support of Y.Z and Z.D, respectively.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Parry EJ. The adulteration of cod liver oil. Lancet. 1904;163:378. [Google Scholar]
- 2.Carter OCS. On the detection of adulterations in oils. Am Philos Soc. 1885;22:296–299. [Google Scholar]
- 3.Jakab A, Heberger K, Forgacs E. Comparative analysis of different plant oils by high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr A. 2002;976:255–263. doi: 10.1016/S0021-9673(02)01233-5. [DOI] [PubMed] [Google Scholar]
- 4.Holčapek M, Jandera P, Zderadička P, Hrubá L. Characterization of triacylglycerol and diacylglycerol composition of plant oils using high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr A. 2003;1010:195–215. doi: 10.1016/S0021-9673(03)01030-6. [DOI] [PubMed] [Google Scholar]
- 5.Park JR, Lee DS. Detection of adulteration in olive oils using triacylglycerols compositions by high temperature gas chromatography. Bull Korean Chem Soc. 2003;24:527–530. doi: 10.5012/bkcs.2003.24.4.527. [DOI] [Google Scholar]
- 6.Lísa M, Holčapek M. Triacylglycerols profiling in plant oils important in food industry, dietetics and cosmetics using high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr A. 2008;1198:115–130. doi: 10.1016/j.chroma.2008.05.037. [DOI] [PubMed] [Google Scholar]
- 7.Christopoulou E, Lazaraki M, Komaitis M, Kaselimis K. Effectiveness of determinations of fatty acids and triglycerides for the detection of adulteration of olive oils with vegetable oils. Food Chem. 2004;84:463–474. doi: 10.1016/S0308-8146(03)00273-5. [DOI] [Google Scholar]
- 8.Ruth SM, Villegas B, Akkermans W, Rozijn M, van der Kamp H, Koot A. Prediction of the identity of fats and oils by their fatty acid, triacylglycerol and volatile compositions using PLS-DA. Food Chem. 2010;118:948–955. doi: 10.1016/j.foodchem.2008.10.047. [DOI] [Google Scholar]
- 9.Woodbury SE, Evershed RP, Rossell JB, Griffith RE, Farnell P. Detection of vegetable oil adulteration using gas-chromatography combustion isotope ratio mass-spectrometry. Anal Chem. 1995;67:2685–2690. doi: 10.1021/ac00111a029. [DOI] [Google Scholar]
- 10.Kelly S, Parker I, Sharman M, Dennis J, Goodall I. Assessing the authenticity of single seed vegetable oils using fatty acid stable carbon isotope ratios (13C/12C) Food Chem. 1997;59:181–186. doi: 10.1016/S0308-8146(96)00286-5. [DOI] [Google Scholar]
- 11.Vigli G, Philippidis A, Spyros A, Dais P. Classification of edible oils by employing 31P and 1H NMR spectroscopy in combination with multivariate statistical analysis. A proposal for the detection of seed oil adulteration in virgin olive oils. J Agric Food Chem. 2003;51:5715–5722. doi: 10.1021/jf030100z. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Lee FSC, Wang XR, He Y. Feasibility study of quantifying and discriminating soybean oil adulteration in camellia oils by attenuated total reflectance MIR and fiber optic diffuse reflectance NIR. Food Chem. 2006;95:529–536. doi: 10.1016/j.foodchem.2005.04.015. [DOI] [Google Scholar]
- 13.Tay A, Singh RK, Krishnan SS, Gore JP. Authentication of olive oil adulterated with vegetable oils using Fourier transform infrared spectroscopy. Lebenson Wiss Technol. 2002;35:99–103. doi: 10.1006/fstl.2001.0864. [DOI] [Google Scholar]
- 14.Gurdeniz G, Ozen B. Detection of adulteration of extra-virgin olive oil by chemometric analysis of mid-infrared spectral data. Food Chem. 2009;116:519–525. doi: 10.1016/j.foodchem.2009.02.068. [DOI] [Google Scholar]
- 15.Tarandjiiska RB, Marekov IN. Precise classification of virgin olive oils with various linoleic acid contents based on triacylglycerol analysis. Anal Chim Acta. 1998;364:83–91. doi: 10.1016/S0003-2670(98)00182-2. [DOI] [Google Scholar]
- 16.Mildner-Szkudlarz S, Jelen HH. The potential of different techniques for volatile compounds analysis coupled with PCA for the detection of the adulteration of olive oil with hazelnut oil. Food Chem. 2008;110:751–761. doi: 10.1016/j.foodchem.2008.02.053. [DOI] [Google Scholar]
- 17.Dourtoglou VG, Dourtoglou Th, Antonopoulos A, Stefanou E, Lalas S, Poulos C. Detection of olive oil adulteration using principal component analysis applied on total and regio FA content. J Am Oil Chem Soc. 2003;80:203–208. doi: 10.1007/s11746-003-0677-1. [DOI] [Google Scholar]
- 18.Jakab A, Jablonkai I, Forgács E. Quantification of the ratio of positional isomer dilinoleoyl-oleoyl glycerols in vegetable oils. Rapid Commun Mass Spectrom. 2003;17:2295–2302. doi: 10.1002/rcm.1193. [DOI] [PubMed] [Google Scholar]
- 19.Goodacre R, Vaidyanathan S, Bianchi G, Kell DB. Metabolic profiling using direct infusion electrospray ionisation mass spectrometry for the characterisation of olive oils. Analyst. 2002;127:1457–1462. doi: 10.1039/b206037j. [DOI] [PubMed] [Google Scholar]
- 20.Ollivier D, Artaud J, Pinatel C, Durbec JP, Guérère M. Differentiation of French virgin olive oil RDOs by sensory characteristics, fatty acid and triacylglycerol compositions and chemometrics. Food Chem. 2006;97:382–393. doi: 10.1016/j.foodchem.2005.04.024. [DOI] [Google Scholar]
- 21.Gómez-Ariza JL, Arias-Borrego A, García-Barrera T, Beltran R. Comparative study of electrospray and photospray ionization sources coupled to quadrupole time-of-flight mass spectrometer for olive oil authentication. Talanta. 2006;70:859–869. doi: 10.1016/j.talanta.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Sikorska E, Górecki T, Khmelinskii IV, Sikorski M, Koziol J. Classification of edible oils using synchronous scanning fluorescence spectroscopy. Food Chem. 2005;89:217–225. doi: 10.1016/j.foodchem.2004.02.028. [DOI] [Google Scholar]
- 23.Mannina L, Patumi M, Fiordiponti P, Emanuele MC, Segre AL. Olive and hazelnut oils: a study by high field 1H-NMR and gas chromatography. Ital J Food Sci. 1999;2:139–149. [Google Scholar]
- 24.Fauhl C, Reniero F, Guillou C. 1H NMR as a tool for the analysis of mixtures of virgin olive oil with oils of different botanical origin. Magn Reson Chem. 2000;38:436–443. doi: 10.1002/1097-458X(200006)38:6<436::AID-MRC672>3.0.CO;2-X. [DOI] [Google Scholar]
- 25.Armenta S, Garrigues S, de la Guardia M. Determination of edible oil parameters by near infrared spectrometry. Anal Chim Acta. 2007;596:330–337. doi: 10.1016/j.aca.2007.06.028. [DOI] [PubMed] [Google Scholar]
- 26.Diaz TG, Merás ID, Casas JS, Franco MFA. Characterisation of virgin olive oils according to its triglycerides and sterols composition by chemometric methods. Food Control. 2005;16:339–347. doi: 10.1016/j.foodcont.2004.03.014. [DOI] [Google Scholar]
- 27.Brodnjak-Vončina D, Kodba ZC, Novič M. Multivariate data analysis in classification of vegetable oils characterized by the content of fatty acids. Chemom Intell Lab Syst. 2005;75:31–43. doi: 10.1016/j.chemolab.2004.04.011. [DOI] [Google Scholar]
- 28.Peña F, Cárdenas S, Gallego M, Valcárcel M. Direct olive oil authentication: Detection of adulteration of olive oil with hazelnut oil by direct coupling of headspace and mass spectrometry, and multivariate regression techniques. J Chromatogr A. 2005;1074:215–221. doi: 10.1016/j.chroma.2005.03.081. [DOI] [PubMed] [Google Scholar]
- 29.Krist S, Stuebiger G, Bail S, Unterweger H. Detection of adulteration of poppy seed oil with sunflower oil based on volatiles and triacylglycerol composition. J Agric Food Chem. 2006;54:6385–6389. doi: 10.1021/jf060500x. [DOI] [PubMed] [Google Scholar]
- 30.Cunha SC, Oliveira MBPP. Discrimination of vegetable oils by triacylglycerols evaluation of profile using HPLC/ELSD. Food Chem. 2006;95:518–524. doi: 10.1016/j.foodchem.2005.03.029. [DOI] [Google Scholar]
- 31.Dugo P, Favoino O, Tranchida PQ, Dugo G, Mondello L. Off-line coupling of non-aqueous reversed-phase and silver ion high-performance liquid chromatography-mass spectrometry for the characterization of rice oil triacylglycerol positional isomers. J Chromatogr A. 2004;1041:135–142. doi: 10.1016/j.chroma.2004.04.063. [DOI] [PubMed] [Google Scholar]
- 32.Asbury GR, Al-Saad K, Siems WF, Hannan RM, Hill HH. Analysis of triacylglycerols and whole oils by matrix-assisted laser desorption/ionization time of flight mass spectrometry. J Am Soc Mass Spectrom. 1999;10:983–991. doi: 10.1016/S1044-0305(99)00063-X. [DOI] [Google Scholar]
- 33.Beermann C, Winterling N, Green A, Möbius M, Schmitt J, Boehm G. Comparison of the structures of triacylglycerols from native and transgenic medium-chain fatty acid-enriched rape seed oil by liquid chromatography–atmospheric pressure chemical ionization ion-trap mass spectrometry (LC–APCI-ITMS) Lipids. 2007;42:383–394. doi: 10.1007/s11745-006-3009-1. [DOI] [PubMed] [Google Scholar]
- 34.Lee DS, Lee ES, Kim HJ, Kim SO, Kim K. Reversed phase liquid chromatographic determination of triacylglycerol composition in sesame oils and the chemometric detection of adulteration. Anal Chim Acta. 2001;429:321–330. doi: 10.1016/S0003-2670(00)01289-7. [DOI] [Google Scholar]
- 35.Webster L, Simpson P, Shanks AM, Moffat CF. The authentication of olive oil on the basis of hydrocarbon concentration and composition. Analyst. 1999;125:97–104. doi: 10.1039/a907036b. [DOI] [Google Scholar]
- 36.Apetrei C, Rodríguez-Méndez ML, de Saja JA. Modified carbon paste electrodes for discrimination of vegetable oils. Sens Actuators B. 2005;111:403–409. doi: 10.1016/j.snb.2005.03.041. [DOI] [Google Scholar]
- 37.Christy AA, Kasemsumran S, Du YP, Ozaki Y. The detection and quantification of adulteration in olive oil by near-infrared spectroscopy and chemometrics. Anal Sci. 2004;20:935–940. doi: 10.2116/analsci.20.935. [DOI] [PubMed] [Google Scholar]
- 38.Zou MQ, Zhang XF, Qi XH, Ma HL, Dong Y, Liu CW, Guo X, Wang H. Rapid authentication of olive oil adulteration by Raman spectrometry. J Agric Food Chem. 2009;57:6001–6006. doi: 10.1021/jf900217s. [DOI] [PubMed] [Google Scholar]
- 39.Møller JKS, Catharino RR, Eberlin MN. Electrospray ionization mass spectrometry fingerprinting of essential oils: Spices from the Labiatae family. Food Chem. 2007;100:1283–1288. doi: 10.1016/j.foodchem.2005.10.013. [DOI] [Google Scholar]
- 40.Obeidat SM, Khanfar MS, Obeidat WM. Classification of edible oils and uncovering adulteration of virgin olive oil using FTIR with the aid of chemometrics. Aust J Basic Appl Sci. 2009;3:2048–2053. [Google Scholar]
- 41.Kurata S, Yamaguchi K, Nagai M. Rapid discrimination of fatty acid composition in fats and oils by electrospray ionization mass spectrometry. Anal Sci. 2005;21:1457–1465. doi: 10.2116/analsci.21.1457. [DOI] [PubMed] [Google Scholar]
- 42.Standal IB, Praël A, McEvoy L, Axelson DE, Aursand M. Discrimination of cod liver oil according to Wild/Farmed and geographical origins by GC and 13C NMR. J Am Oil Chem Soc. 2008;85:105–112. doi: 10.1007/s11746-007-1174-x. [DOI] [Google Scholar]
- 43.Rohman A, Man YBC. Analysis of cod-liver oil adulteration using Fourier transform infrared (FTIR) spectroscopy. J Am Oil Chem Soc. 2009;86:1149–1153. doi: 10.1007/s11746-009-1453-9. [DOI] [Google Scholar]
- 44.Cozzolino D, Murray I, Chree A, Scaife JR. Multivariate determination of free fatty acids and moisture in fish oils by partial least-squares regression and near-infrared spectroscopy. LWT-Food Sci Technol. 2005;38:821–828. doi: 10.1016/j.lwt.2004.10.007. [DOI] [Google Scholar]
- 45.Aursand M, Jørgensen L, Grasdalen H. Positional distribution of ω3 fatty acids in marine lipid triacylglycerols by high-resolution 13C nuclear magnetic resonance spectroscopy. J Am Oil Chem Soc. 1995;72:293–297. doi: 10.1007/BF02541085. [DOI] [Google Scholar]
- 46.Standal IB, Axelson DE, Aursand M. Differentiation of fish oils according to species by 13C-NMR regiospecific analyses of triacyglycerols. J Am Chem Soc. 2009;86:401–407. doi: 10.1007/s11746-009-1370-y. [DOI] [Google Scholar]
- 47.Martinez I, Standal IB, Axelson DE, Finstad B, Aursand M. Identification of the farm origin of salmon by fatty acid and HR 13C NMR profiling. Food Chem. 2009;116:766–773. doi: 10.1016/j.foodchem.2009.03.026. [DOI] [Google Scholar]
- 48.Araujo P, Nguyen TT, Frøyland L, Wang JD, Kang JX. Evaluation of a rapid method for the quantitative analysis of fatty acids in various matrices. J Chromatogr A. 2008;1212:106–113. doi: 10.1016/j.chroma.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]