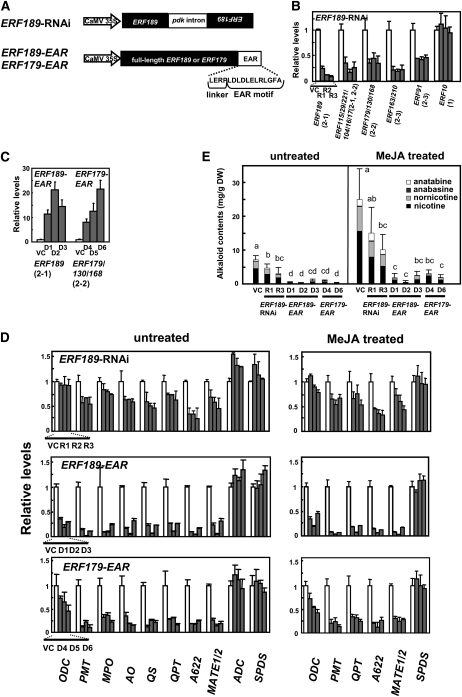
Figure 5.
Downregulation or Dominant Repression of NIC2-Locus ERF Genes Inhibits Nicotine Biosynthesis in Transgenic Tobacco Roots.
Transcript levels are shown relative to empty vector control values, and error bars indicate the sd for three biological replicates. Clade numbers are indicated in parentheses.
(A) Schematic diagrams of the ERF189-RNAi, ERF189-EAR, and ERF179-EAR constructs. The ERF189-RNAi construct contains an intron of PDK (pyruvate orthophosphate dikinase from Flaveria trinerva) and two flanking ERF189 fragments in opposite orientations. ERF189-EAR and ERF179-EAR are fusion proteins with an EAR motif attached to the C termini of ERF189 and ERF179 via a short linker.
(B) Transcript levels of ERF genes in three ERF189 lines (R1 to R3; gray bars) and a vector control line (VC; white bars). Expression levels of clade 2 ERFs and clade 1 ERF10 were analyzed by qRT-PCR.
(C) Transcript levels of ERF genes in three ERF189-EAR lines (D1 to D3), three ERF179-EAR lines (D4 to D6), and a vector control line (VC). qRT-PCR was used to measure expression levels of both the endogenous ERF genes and the introduced transgenes.
(D) Transcript levels of tobacco metabolic enzyme genes. Tobacco hairy roots were cultured in the absence or presence of 100 μM MeJA for 24 h and analyzed for expression of the structural genes of nicotine biosynthesis (ODC, MPO, PMT, AO, QS, QPT, A622, and MATE1/2) and the control genes (ADC and SPDS) by qRT-PCR. In the VC line, MeJA increased the transcript levels as follows: ODC, 5.6-fold; PMT, 9.5-fold; QPT, 8.4-fold; A622, 9.2-fold; MATE1/2, 7.2-fold; and SPDS, 1.1-fold.
(E) Alkaloid levels in the transgenic root lines. In one experiment, tobacco hairy roots were treated with 100 μM MeJA for 3 d. Significant differences among the lines were determined at P < 0.05 by one-way analysis of variance, followed by the Tukey-Kramer test, and are indicated by different letters. DW, dry weight.