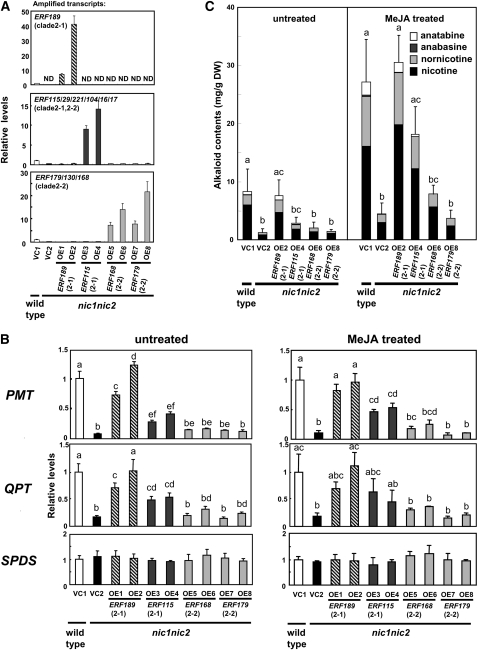
Figure 6.
Complementation of nic1 nic2 Roots by Overexpression of ERF Genes.
Transcript levels are shown relative to the values for the wild type transformed with the empty vector (VC1), and error bars indicate the sd for three biological replicates. Significant differences among the lines were determined at P < 0.05 by one-way ANOVA, followed by the Tukey-Kramer test, and are indicated by different letters. Clade numbers are indicated in parentheses.
(A) Overexpression of ERF189, ERF115, ERF168, or ERF179 in nic1 nic2 tobacco hairy roots. The vector control lines VC1 and VC2 were generated by transforming wild-type and nic1 nic2 plants, respectively, with an empty vector. Transcript levels include both the endogenous genes and the transgenes. ND, not detected.
(B) Relative expression levels of PMT, QPT, and SPDS in the transgenic root lines. For MeJA treatment, the hairy roots were elicited with 100 μM MeJA for 24 h. In the VC1 line, MeJA increased the transcript levels as follows: PMT, 8.7-fold; QPT, 7.9-fold; and SPDS, 1.2-fold.
(C) Alkaloid levels in the transgenic root lines. The right graph shows data from tobacco hairy roots treated with 100 μM MeJA for 3 d. DW, dry weight.