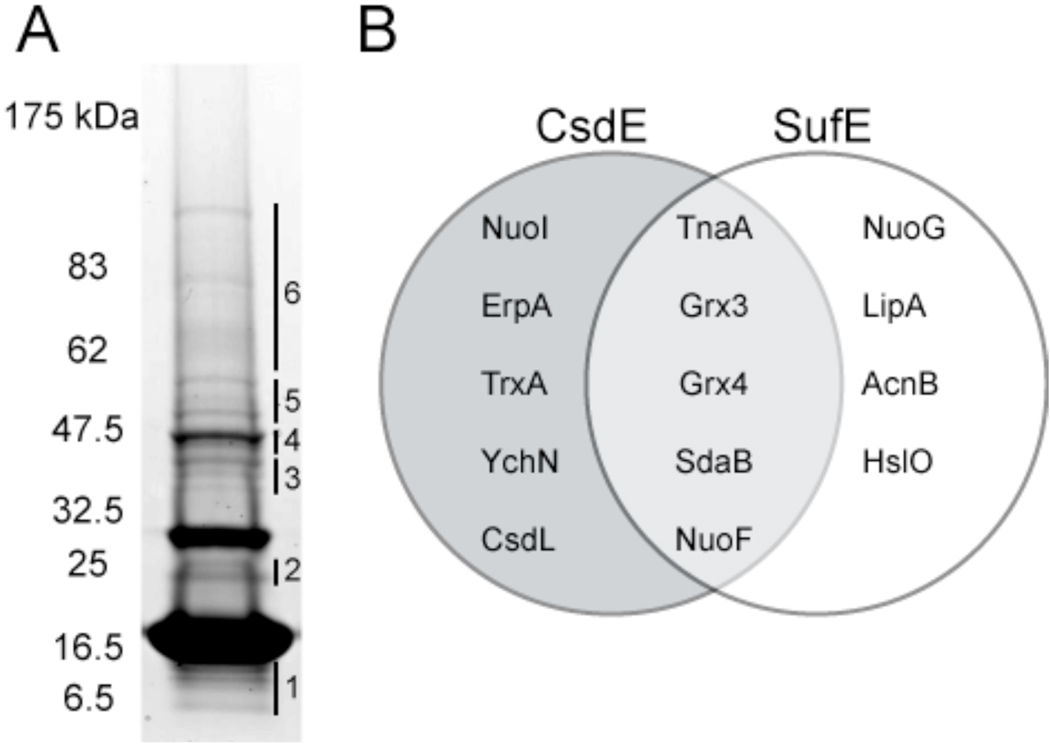
Figure 4. Identification of CsdE interactome by LC-MS/MS mass spectrometry and comparison with SufE interactome.
A, TCA-prepared protein extracts from cells expressing CsdAE-(His)6 were purified by Ni2+ affinity chromatography, resolved by reducing SDS-PAGE, and stained with CBB. The CsdE monomer is observed at ~16.5 kDa. The gel was subsectioned and in-gel proteolysis followed by LC-MS/MS was performed on each section. CsdA was identified in section #4. SI Tables S2 and S3 list all 116 proteins that were identified with a minimum of 2 peptides and 95% confidence in the subsections. Table 1 presents the proteins of particular interest that were identified. B, Comparison between the CsdE and SufE interactomes (SufE interactome presented in Bolstad and Wood, 2010, submitted). Proteins were identified by LC-MS/MS. Only those proteins that represent interactions of predicted significance are shown.