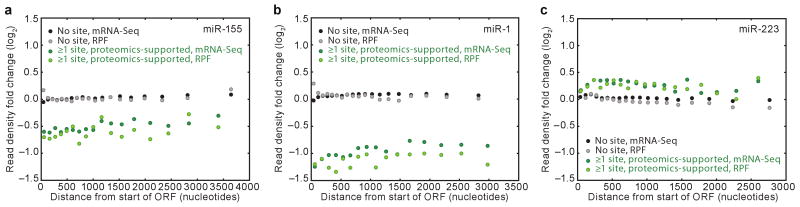
Figure 4. Ribosome and mRNA changes were uniform along the length of the ORFs.
a, Ribosome and mRNA changes along the length of ORFs after introducing miR-155. mRNA segments of quantified genes were binned based on their distance from the first nucleotide of the start codon, with the boundaries of the segments chosen such that each bin contained the same number of nucleotides (Supplementary Fig. 8b). Binning was done separately for mRNAs with no miR-155 site and proteomics-supported miR-155 targets. Fold changes in RPFs and mRNA-Seq tags mapping to each bin were then plotted with respect to the median distance of the central nucleotide of each segment from the first nucleotide of the start codon. Changes in RPFs and mRNA-Seq tags for mRNAs with no site (grey and black, respectively) and for proteomic-supported targets (light and dark green, respectively) are shown. Only bins with read contribution from ≥20 genes are shown (see Supplementary Fig. 8b). The ANCOVA test for systematic change across the ORF length was performed by first calculating the differences between RPF changes and mRNA-Seq changes for each group of genes, fitting lines through these changes in translational efficiency, then testing for a difference between the resulting slopes. b, As in panel a, but plotting results for the miR-1 experiment. c, As in panel a, but plotting results for the miR-223 experiment.