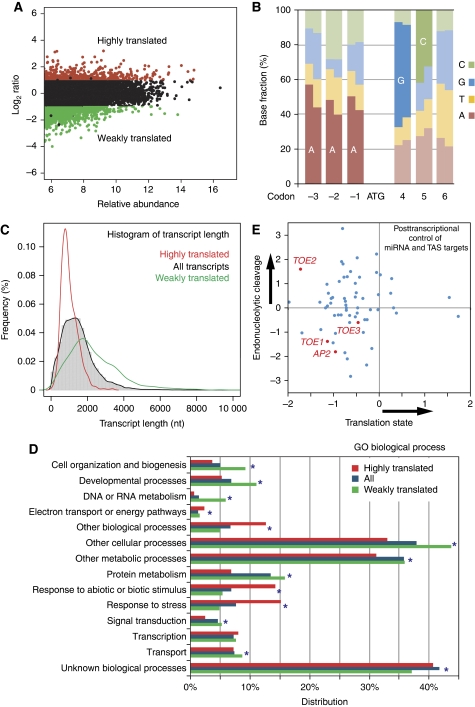
Figure 4.
Translational regulation during flower development. (A) M-A plot in which log2-transformed ratios of translating versus total mRNA signal for stage 4 flowers are plotted against log-intensity averages. Transcripts with statistically different levels in the translating and total RNA populations (P<0.001) and a ratio above two are considered as highly translated and are shown in red. Transcripts with statistically different levels and ratio below 0.5 were considered as weakly translated and are shown in green. Plants expressing HF–RPL18 under the RPL18 promoter were used for translatome profiling. (B) Biases around the start codon shown as the fractions of bases. At each base (rectangle), the left half represents the fraction of each base in highly translated genes and the right half represents the fraction of each base in weakly translated genes. Enriched bases are labeled. (C) Enhanced translation for short transcripts. A transcript length distribution of all expressed genes is shown by the histogram and the black line, while red and green lines show distributions of highly and weakly translated transcripts, respectively. (D) Relationship between gene function and translation levels. All genes detected as expressed for ‘Biological Process’ categories (Total) are compared with highly translated (red) and weakly translated (green) transcripts. ‘Distribution’ refers to the percentages of genes annotated to descriptive terms in a particular GO category divided by all genes. Categories with FDR corrected hypergeometric test P<0.001 are marked with stars. (E) Estimated regulation modes of miRNA target genes. Scatterplot of uncapping levels (endonucleolytic cleavage) of miRNA and ta-siRNA (TSA) target mRNAs versus translation efficiency (translation inhibition) of the same mRNAs in stage 4 flower tissues. Targets of miR172, including AP2 and AP2-like mRNAs are highlighted in red.