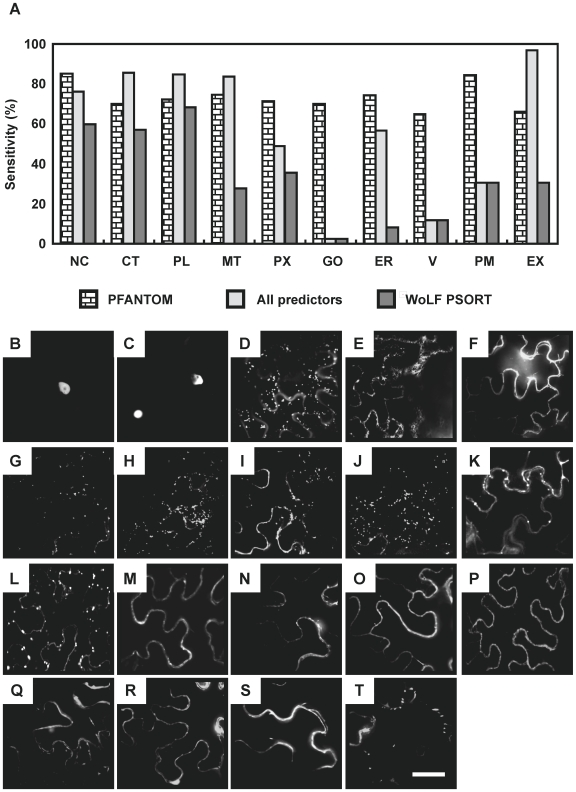
Figure 3. Validation of the PFANTOM method for plant subcellular localizations.
(A) Comparison of the prediction performance of Pfam-based prediction (PFANTOM), all predictors by SUBA database, and WoLF PSORT. NC, nucleus; MT, mitochondrion; V, vacuole; PX, peroxisome; ER, endoplasmic reticulum; GO, Golgi apparatus; CT, cytosol; PM, plasma membrane; PL, plastid; EX, extracellular. (B-T) Subcellular localization of transiently expressed YFP-fusion proteins in N. benthamiana. (B) bHLH (AT5G48560); (C) SND1 (AT1G32770); (D) Golgi marker (STtmd-GFP); (E) ER marker (GFP-HDEL); (F) Plasma membrane marker (pm-rk); (G) PGSIP1 (AT3G18660); (H) PGSIP3 (AT4G33330); (I) TBL3 (AT5G01360, belonging to DUF231); (J) Unknown protein (AT2G38320, belonging to DUF231); (K) CTL2 (AT3G16920); (L) FLA11 (AT5G03170); (M) FLA12 (AT5G60490); (N) COBL4 (AT5G15630, IRX6); (O) LRR protein (AT1G67510); (P) NCRK (AT2G28250); (Q) RIC2 (AT1G27380); (R) RIC4 (AT5G16490); (S) ROP7 (AT5G45970); (T) ROPGEF4 (AT2G45890). A summary of the localization experiments is shown in Table 2. Scale bar = 20 µm.