Figure 4. TNF-α promotes binding of NF-κB subunits p50 and p65 to the NHE2 promoter region.
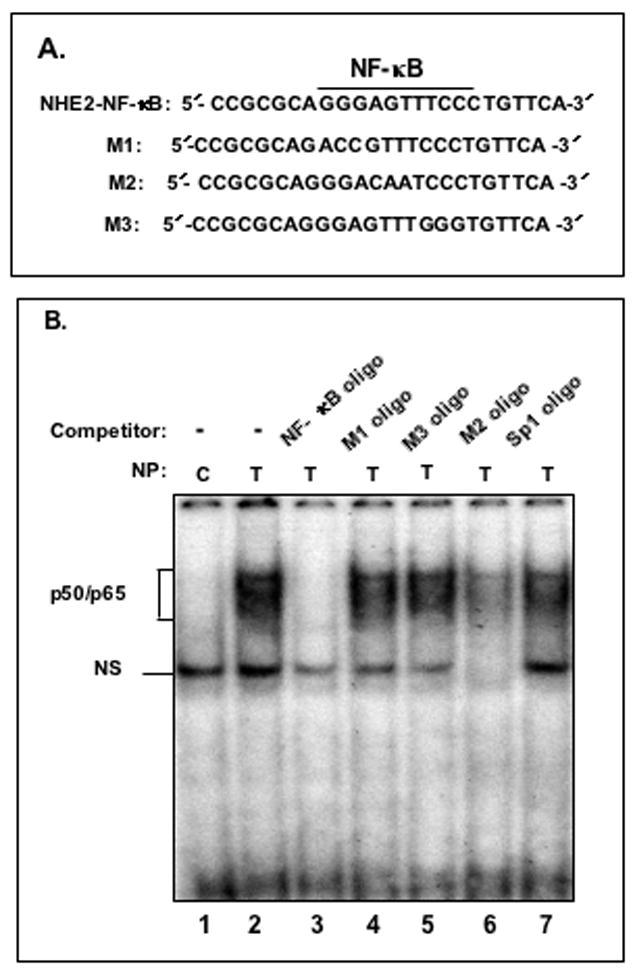
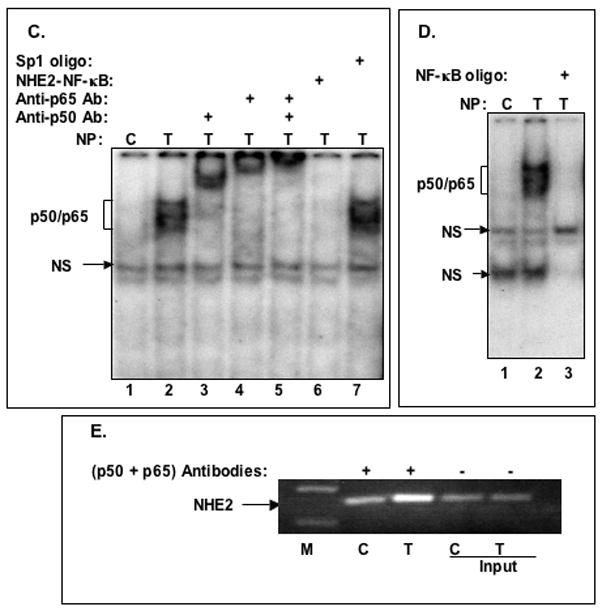
A: Oligonucleotides used in GMSA. B: Binding of nuclear proteins in TNF-α treated cells to 32P-labeled NHE2-NF-κB probe. Nuclear proteins (5 μg) from untreated (lane 1) or TNF-α treated cells (lanes 2–7) were allowed to interact with the probe in the presence of molar excess of cold competitor oligonucleotides NF-κB consensus sequence, M1, M3, M2 and Sp1 consensus sequence (lanes 3–7, respectively). The unlabeled competitor probes were incubated with the nuclear proteins for 10 minutes prior to addition of radiolabeled NHE2-NFκB probe (50,000 cpm). C: p50 and p65 interact with the NHE2 promoter. NF-κB activation by TNF-α was determined in nuclear proteins from untreated (control) and cells treated with TNF-α for 1. The identities of the proteins present in these complexes were established by supershift assays. Incubation of anti-p50 and anti-p65 antibodies individually (lanes 3 and 4, respectively) or simultaneously (lane 5) resulted in the formation of slow migrating supershifted bands (SS), suggesting the presence of both proteins in the DNA-protein complexes. A 100-molar excess of unlabeled NHE2-NF-κB probe and Sp1 oligonucleotide were used in competition assays (lanes 6 and 7). D: GMSA with NF-κB consensus oligonucleotide used as a positive control. E: ChIP Assay. ChIP assay was performed as described in Materials and Methods. Immunoprecipitated DNA was analyzed by PCR using NHE2 promoter-specific primers. To verify that an equivalent amount of chromatin was used in the immuno-precipitations, an input chromatin was amplified with the same primers as control. C and T, indicate the nuclear proteins from control or TNF-α treated C2BBe1 cells, respectively. NS indicates non-specific binding. M, DNA marker.
