Figure 1.
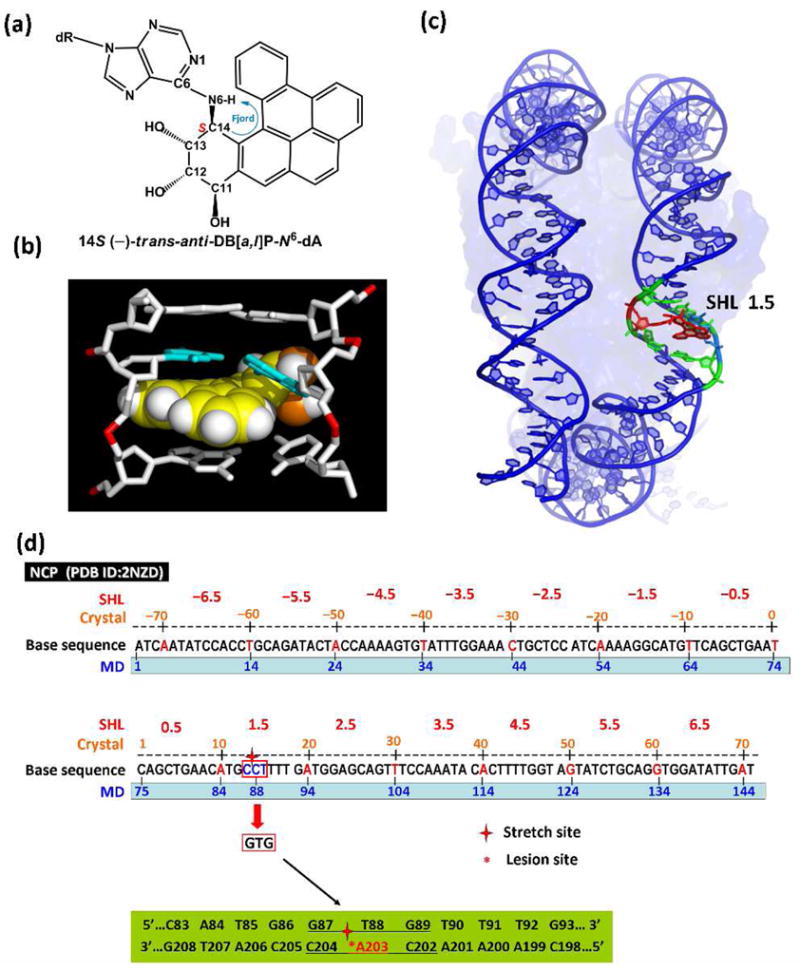
(a) chemical structure of the 14S (−)-trans-anti-DB[a,l]P-N6-dA adduct, and intercalation models for (b) the uncomplexed duplex (see Supporting Information) and (c) the nucleosome following 65-ns of MD simulations. See also Movie S1 Supporting Information. (d) Base sequence contexts of the NCP with a 145-mer DNA duplex (PDB ID : 2NZD) (17) with numbering schemes in the crystal structure and MD simulations. The sequences surrounding the stretch site at SHL 1.5, circled in red, were remodeled to the …GTG… : …CAC… duplex (…G87-T88-G89… : …C204-A203-C202…). The 11-mer centered at the lesion-modified base pair A203* : T88 is highlighted in green.
