Abstract
Huntington's disease (HD) is a currently incurable neurodegenerative disease caused by the expansion of a CAG trinucleotide repeat within the huntingtin (HTT) gene. Therapeutic approaches include selectively inhibiting the expression of the mutated HTT allele while conserving function of the normal allele. We have evaluated a series of antisense oligonucleotides (ASOs) targeted to the expanded CAG repeat within HTT mRNA for their ability to selectively inhibit expression of mutant HTT protein. Several ASOs incorporating a variety of modifications, including bridged nucleic acids and phosphorothioate internucleotide linkages, exhibited allele-selective silencing in patient-derived fibroblasts. Allele-selective ASOs did not affect the expression of other CAG repeat-containing genes and selectivity was observed in cell lines containing minimal CAG repeat lengths representative of most HD patients. Allele-selective ASOs left HTT mRNA intact and did not support ribonuclease H activity in vitro. We observed cooperative binding of multiple ASO molecules to CAG repeat-containing HTT mRNA transcripts in vitro. These results are consistent with a mechanism involving inhibition at the level of translation. ASOs targeted to the CAG repeat of HTT provide a starting point for the development of oligonucleotide-based therapeutics that can inhibit gene expression with allelic discrimination in patients with HD.
Keywords: Locked nucleic acid (LNA), allele-selective inhibition, Huntington's Disease, huntingtin, antisense oligonucleotide, CAG repeat, triplet repeat
Expansion of unstable trinucleotide repeats is responsible for at least 16 inherited neurological disorders (1). Although Huntington's disease is one of the most well known trinucleotide repeat diseases, it remains incurable and insufficiently understood. HD has a prevalence of 1 in 10,000–15,000 in populations of European descent and is characterized by adult onset and progressive neurodegeneration (2). Symptoms include chorea, dystonia and cognitive or psychiatric disturbances. These symptoms gradually worsen until death 10–20 years after disease onset (2, 3).
The expansion of a CAG repeat in exon 1 of the huntingtin (HTT) gene leads to the autosomal dominant disease (3, 4). Expansion beyond 36 repeats is associated with HD and full disease penetrance is observed for repeat lengths >41 (5–8). The CAG repeat encodes a polyglutamine (poly-Q) tract at the N-terminus of the HTT protein and repeat length is inversely correlated with age at disease onset in patients with HD (2, 7). The expanded poly-Q tract in HTT confers a gain of function, namely oligomerization of mutant HTT poly-Q fragments, disables efficient protein degradation pathways, and may interfere with normal protein functions (2, 4, 9, 10). Aggregation of poly-Q containing HTT fragments eventually induces the formation of plaques in the brains of patients with HD (11). Cells in the central nervous system and brain are sensitive to these changes, especially the cortico-striatal neurons of the striatum which suffer the greatest degeneration (2, 9, 11).
Therapeutic options for HD include small molecule drugs like haloperidol, tetrabenazine, clonazepam, fluoxitine, and sertraline that are designed to control the phenotypic manifestations of the disease (12–15). While these drugs can improve quality of life for patients with HD, they are not expected to significantly reverse or alter disease progression or increase life expectancy, nor do they address the underlying molecular mechanisms of the disease (12).
HTT is a large protein (348 kDa) predicted to interact with over 180 proteins and has been reported to influence several pathways, including transcription, apoptosis, mitochondrial function and energy metabolism, tumor suppression, vesicular and neurotransmitter release and axonal transport (4, 16–19). Drug development has targeted several cellular pathways, including co-enzyme Q10 analogs for mitochondrial dysfunction and farnesyltransferase inhibitors to upregulate autophagy (20, 21). Therapeutic intervention is complicated because targeting a single pathway may only partially alleviate the disease.
One direct route to treating HD would be reduction or elimination of the causative agent, the mutant protein itself. HTT is an essential gene that is required in embryogenesis, neurogenesis and normal adult function (22, 23) and evidence suggests that 30–40% expression of wild-type HTT is necessary to support normal development and function (21, 23–25). Thus, to maintain sufficient levels of normal HTT protein, and because almost all patients have one normal and one mutant HTT gene (4, 7), potential therapeutic strategies include reducing expression of the mutant HTT allele (26).
One strategy for allele-selective inhibition of HTT expression is development of small interfering RNAs (siRNAs) that target single-nucleotide polymorphisms (SNPs) or deletion polymorphisms (27–31). Recent reports suggest that a small number of siRNAs targeting different SNP sites could potentially treat >75% of patients with HD (31, 32). However, the need to develop a collection of related drugs would complicate clinical development. Thus, while SNP-targeted siRNAs are an important approach for developing selective agents, there remains a need for alternate strategies.
Single-stranded complementary oligomers containing peptide nucleic acid (PNA) or locked nucleic acid (LNA) can selectively inhibit expression of the mutant HTT allele (33–35). A similar antisense approach has been described recently in which complementary morpholino oligomers block the binding of muscle-blind like 1 (MBNL-1) to expanded CUG repeats and affect the pathogenesis of myotonic dystrophy (36). Antisense oligonucleotides (ASOs) contain a single nucleic acid strand that can be heavily modified to enhance stability, target binding and biodistribution. ASOs have been extensively investigated in the clinic as a therapeutic approach (26, 37, 38) and promising Phase III data has been recently presented for Mipomersen, an ASO that reduces expression of apolipoprotein B (39–41).
Here we test a broad spectrum of modified ASOs targeted to the CAG repeat within HTT mRNA and identify several that achieve allele-selective inhibition. ASOs targeting expanded trinucleotide repeat regions of mRNA may represent a route to allele-selective therapeutics for treatment of HD and other repeat expansion diseases.
MATERIALS AND METHODS
Oligonucleotides
ASOs were synthesized and purified by Isis Pharmaceuticals, Sigma Custom Products, Glen Research Corporation and the Damha laboratory. Unmodified DNA and RNA were purchased from Integrated DNA Technologies, Inc.
Thermal Denaturation by UV Melt Analysis
Thermal denaturation analysis of ASOs or ASO:RNA duplexes was carried out using a CARY Varian model 3 UV-Vis spectrophotometer. Absorbance was monitored at 260 nm in a 1 cm quartz cuvette. ASO (0.8 μM) or ASO annealed to complementary RNA (0.8 μM) in 1× Dulbecco's phosphate-buffered saline (Sigma Aldrich) was annealed and melted three times from 18°C to 99°C at a ramp rate of 2°C/min. To evaluate the concentration dependence of Tm, 0.1 cm quartz cuvettes were used with variable concentrations of oligonucleotide ranging from 0.5 to 50.0 μM. Absorbance was collected at one reading per 1°C. Tm was calculated using CARY WinUV Thermal Application software using a baseline fitting method.
Thermal Denaturation by Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) was performed on a MicroCal VP-DSC capillary cell microcalorimeter. ASOs or ASO:RNA duplexes at 10 or 20 μM in degassed 1× Dulbecco's phosphate-buffered saline (Sigma Aldrich) were first heated to 95°C for 5 min then cooled to room temperature on the bench top to allow duplex annealing. Samples were scanned under pressure from 35°C to 125°C at a scan rate of 90°C/hr with 2 rescans. The peak change in heat capacity (Cp) was determined using the Origin 7.0 with DSC Data Analysis (MicroCal) software. The temperature corresponding to the maximum peak in Cp of the ASO or ASO:RNA duplex was taken as the Tm. Replicate scans were used to calculate the average Tm and standard deviation. Origin 7.0 with DSC Data Analysis software was also used to fit DSC scans to a 2-state unfolding model and calculate enthalpy.
Cell Culture and Transfection
Patient-derived fibroblast cell lines GM04281 (69 CAG repeat mutant allele), GM04717 (41 CAG repeat mutant allele), and GM04719 (44 CAG repeat mutant allele) were obtained from the Coriell Institute. Cells were maintained at 37°C and 5% CO2 in MEM (Sigma) supplemented with 10% FBS (Sigma) and 0.5% MEM nonessential amino acids (Sigma). Cells were plated in 6-well dishes at 60,000 cells/well in supplemented MEM 2 days before transfection. Stock solutions of modified ASOs were heated at 65°C for 5 min prior to use to dissolve any aggregation. Modified ASOs were transfected into cells using RNAiMAX (Invitrogen) according to the manufacturer's instructions. The appropriate amount of lipid (2 μl for 125 pmol ASO) was added to OptiMEM containing oligonucleotides and the oligonucleotide-lipid mixture (250 μl) was incubated for 20 min. OptiMEM was added to the mixture to a final volume of 1.25 ml and then added to cells. Media was exchanged 1 day after transfection with fresh supplemented MEM. Cells were washed with phosphate-buffered saline and harvested 4 days after transfection for protein analysis or at 3 days for RNA analysis.
Western blot analysis
Cells were harvested with trypsin-EDTA solution (Invitrogen) and lysed. The protein concentration in each sample was quantified with micro-bicinchoninic acid (micro-BCA) assay (Thermo Scientific). SDS-PAGE (separating gel: 5% acrylamidebisacrylamide [50:1], 450 mM Tris-acetate pH 8.8; stacking gel: 4% acrylamide-bisacrylamide [50:1], 150 mM Tris-acetate pH 6.8) was used to separate wild-type and mutant HTT proteins. Gels were run at 30 mA per gel for 5 hr in Novex Tris-Acetate SDS Running Buffer (Invitrogen). For separation of HTT variants containing shorter CAG repeats, gels were run for 6–7 hr. The electrophoresis apparatus was placed in a 15°C water bath to prevent overheating. In parallel with analysis for HTT expression, samples were analyzed for β-actin expression by SDS-PAGE (7.5% acrylamide pre-cast gels; Bio-Rad) to ensure even loading of protein in all lanes. These gels were run at 80 V for 15 min followed by 100 V for 1 hr in 1× TGS buffer (Bio-Rad).
After electrophoresis, proteins were transferred to membrane (Hybond-C Extra; GE Healthcare Bio-Sciences). Primary antibodies specific for HTT (MAB2166, Chemicon) and β-actin (Sigma) protein were obtained and used at 1:10,000 dilutions. HRP conjugate anti-mouse secondary antibody (1:10,000, Jackson ImmunoResearch Laboratories) was used for visualizing proteins using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Detection of TBP and FOXP2 was performed similar to that of a previous description (33). Briefly, samples were run on 7.5% precast gels (Bio-Rad), transferred to membrane and probed with a 1:5000 or 1:1000 dilution of anti-TBP (Sigma) or anti-FOXP2 (Abcam), respectively, at 4°C overnight. HRP conjugate anti-mouse or anti-rabbit (1:5000) (Jackson ImmunoResearch Laboratories) was used to visualize TBP and FOXP2, respectively, with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific). Protein bands were quantified from autoradiographs using ImageJ software. Percentage of inhibition was calculated as a relative value to control samples.
Each data plot from dose response experiments for inhibition of HTT was fit to the following model equation: y = 100(1−xm/(nm+xm)) using Prism 4.0 (GraphPad), where y is percent expression of HTT protein and x is concentration of ASO. Both m and n are fitting parameters, where n is taken as the IC50 value. The IC50 values were calculated from individual dose responses fit to the above equation and then reported as the mean and standard error of the mean of three or more biological replicates.
Quantitative PCR (qPCR)
Total RNA from fibroblast cells was extracted using TRIzol (Invitrogen) 3 days after transfection. Two micrograms of RNA was treated with 2 units of DNase I (Worthington Biochemical Corp.) for 10 min at 25°C. RNA was reverse transcribed using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Quantitative PCR was performed on a BioRad CFX96 Real Time System using iTaq SYBR Green Supermix with ROX (Bio-Rad). Data were normalized relative to levels of GAPDH mRNA. Primers specific for the HTT exon 64–65 boundary were used: forward primer, 5′-CGACAGCGAGTCAGTGAATG–3′; reverse primer, 5′-ACCACTCTGGCTTCACAAGG–3′. Primers specific for GAPDH were obtained from Applied Biosystems. Experiments were performed in biological triplicate and error reported as standard deviation.
Cloning and in vitro transcription of 5′ truncated HTT mRNA transcripts
Sequence corresponding to the 5′ end of HTT mRNA was cloned from GM04281 fibroblasts. Total RNA was extracted with TRIzol (Invitrogen), DNase I-treated and reverse transcribed with random primers using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Complementary DNA was amplified by PCR with HotStarTaq DNA Polymerase (Qiagen) following the manufacturer's protocol and using the following primers: forward primer, 5′-ATGGCGACCCTGGAAAAG-3′; reverse primer, 5′-GGCTGAGGAAGCTGAGGAG-3′. PCR products were gel-purified and cloned into the pCR 4-TOPO plasmid using the TOPO-TA Cloning Kit (Invitrogen) following the manufacturer's recommended protocol. The cloning of the 5′ cDNA sequence of HTT mRNA was verified by sequencing.
Plasmids were linearized with Pme I restriction enzyme and used for the production of REP69 or REP17 RNAs by in vitro run-off transcription using an Ampliscribe T7-Flash Transcription Kit (Epicentre). RNAs were extracted with pH 4.3 phenol and 24:1 chloroform:isoamyl alcohol. One volume of gel purification buffer (1× TBE, 90% formamide) was added and the RNA solution boiled for 5 min. RNA samples were resolved on 6% denaturing polyacrylamide gels (7 M urea, 1× TBE) and the bands visualized by lightly staining with methylene blue before being cut out. RNA was extracted by crush and soak elution with RNA elution buffer (20 mM Tris, pH 7.4, 5 mM EDTA, 0.3 M sodium acetate, 0.1% SDS) then precipated with two volumes of ethanol.
Electrophoretic Mobility Shift Assays (EMSA)
Twenty pmols of REP69 or REP17 RNA was dephosphorylated in a 40 μL reaction containing 10 U of calf intestinal phosphatase (CIP) (New England Biolabs), 1× CIP buffer and 20 U of SUPERase-In RNase Inhibitor (Ambion) for 15 min at 37°C then 15 min at 55°C. RNA was extracted with pH 4.3 phenol and 24:1 chloroform:isoamyl alcohol then precipitated with 0.3 M sodium acetate and 70% ethanol. CIP-treated RNA was 5′-end labeled in a 25 μL reaction containing 20 U of T4 polynucleotide kinase (T4 PNK) (New England Biolabs), 1× PNK buffer, 20 U of SUPERase-In and 3 μL of [γ]-32P ATP (7000 Ci/mmol) (MP Biomedicals) then incubated at 37°C for 1.5 hr. Radiolabeled RNA was extracted with pH 4.3 phenol and 24:1 chloroform:isoamyl alcohol. RNA was gel-purified as described above for unlabeled RNA, except that the RNA bands were visualized by a phosphorimager, and radioactivity quantified by liquid scintillation counting.
To evaluate ASO binding to structured REP17 and REP69 RNAs, electrophoretic mobility shift assays (EMSA) were performed. Radiolabeled RNA was refolded in 1× EMSA buffer (25 mM Phosphate buffer, pH 7.0, 0.2 mM EDTA, 0.15 M NaCl, 1 mM MgCl2) by heating to 98°C then cooling to 25°C at a rate of 0.1°C/sec. Increasing concentrations of ASO were mixed with the refolded RNA (80 fmols, 20,000 cpm) in a 10 μL reaction containing 1× EMSA buffer, 5 U SUPERase-In, and 10 μg yeast tRNA and incubated at 37°C for 15 min. Reactions were moved to room temperature, 2.5 μL of phosphate buffer dye (0.25 M phosphate buffer, pH 7.0, 50% glycerol, 0.02% bromophenol blue, 0.02% xylene cyanol) added, and then loaded onto a 0.75 mm thick native polyacrylamide gel (25 mM phosphate, pH 7.0, 6% 19:1 acrylamide:bisacrylamide, 2% glycerol). Gels were run with cooling for 5–6 hr at 150 volts then dried to 3M Whatman paper and exposed to phosphorimager cassettes for visualization.
Shifted RNA was quantified with ImageJ software and fit to a Hill equation: y = (xn)/(Kdn+xn) using Prism 4.0 (GraphPad), where y is the fraction of RNA shifted and x is the concentration of ASO. Both Kd and n are fitting parameters, where Kd is taken as the dissociation constant and n is the Hill coefficient.
In Vitro RNase H Activity Assay
Radiolabeled REP69 RNA (0.1 pmol, 20,000 cpm) was incubated with ASO (10 pmol) in 1× EMSA buffer in a 9.5 μL reaction for 5 min at 37°C. RNase H1 enzyme from Escherichia coli (1 U) (Invitrogen) was added for a final reaction volume of 10 μL and incubated for 10 min at 37°C. Reactions were stopped by the addition of 9 volumes of 2% LiClO4 in acetone. Reactions were spun down, pellets washed with acetone and resuspended in gel purification buffer, boiled for 5 min, and then resolved on 5% denaturing polyacrylamide gels supplemented with 2% glycerol. Gels were run at 250 volts for 2.5 hr, dried to 3M Whatman paper and visualized by phosphorimager.
RESULTS
Strategy for Designing and Testing Modified Oligonucleotides
We designed modified antisense oligonucleotides (ASOs) to optimize allele-selective inhibition and gain further insights into mechanism. Our lead compound was LNA(T) (33, 34). LNA(T) is a 19 nucleotide long oligonucleotide consisting of repeating GCT sequence with 3′ and 5′ terminal guanines. LNA(T) targets the CAG repeat sequence in exon 1 of HTT mRNA and contains deoxyribonucleic acid (DNA) nucleotides mixed with LNA nucleotides at every third base, in this case at every thymine residue (Figure 1A). Thermodynamic effects of the LNA are largely propagated to the nearest neighboring 5′ and 3′ nucleotides (42). Thus, for a G-C rich 19 base oligonucleotide, this configuration is preferred because it produces high affinity binding and minimizes manufacturing costs.
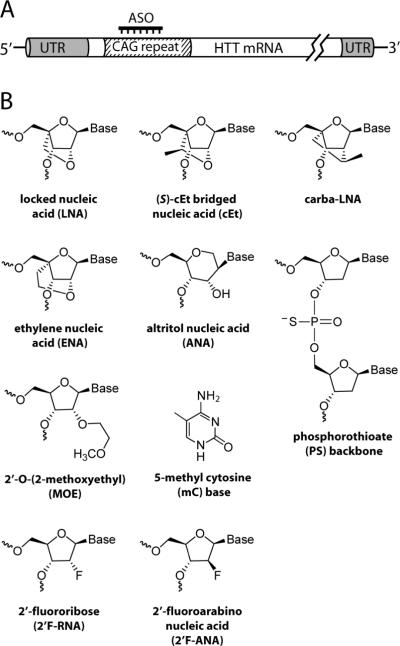
Figure 1. Strategy for targeting the expanded CAG repeat region of mutant HTT mRNA to elicit allele-selective inhibition.
(A) Schematic of the HTT mRNA and the CAG repeat region that is targeted with complementary CTG-repeat ASOs. UTR, untranslated region. (B) Chemically modified base, backbone and nucleosides incorporated into ASOs tested for allele-selective inhibition of HTT expression.
LNA is a member of a class of modifications called bridged nucleic acid (BNA) (43, 44). BNA is characterized by a covalent linkage that locks the conformation of the ribose ring in a C3′-endo (northern) sugar pucker. For LNA, the bridge is composed of a methylene between the 2′-O and the 4′-C positions. LNA enhances backbone pre-organization and base stacking to increase hybridization and thermal stability (Figure 1B) (43–46).
Three other BNA modifications were tested: 2′-O, 4′-C-ethylene-bridged nucleic acid (ENA), 2′, 4′-constrained ethyl nucleic acid called S-cEt (cEt), and 2′-4′-carbocyclic LNA (carba-LNA) (Figure 1B). These modifications enhance the affinity of binding and protect the oligonucleotide against nuclease digestion. ENA provides a more flexible bridge than LNA (47, 48). The cEt modification has been shown to possess a favorable toxicity profile in animals (49, 50). Carba-LNA retains LNA-like properties but is chemically unique in that it lacks oxygen on the 2′ carbon (51). We also tested altritol nucleic acid (ANA) consisting of a six-membered D-altritol sugar with a nucleobase attached at the 2′-position and a hydroxyl group at the 3′-position. ANA rigidifies the base conformation, improves thermal stability, and reduces digestion by nuclease (52). A practical advantage for all BNA nucleotides is that they can be substituted within RNA or DNA oligomers, simplifying synthesis and allowing the properties of the parent oligomer to be fine-tuned for specific applications.
We also examined substitution at the 2′ position of the ribose with fluorine in either stereoisomer orientation (2′F-RNA or 2′F-ANA) or 2′-O-(2-methoxyethyl) (MOE) (Figure 1B). These 2′ modifications improve nucleic acid hybridization and duplex stability (53–55). 2′F-ANA is known to support RNase H cleavage with efficiencies that vary depending on the location of substitution (55, 56). Some oligomers were “gapmer” designs that incorporate modifications near the termini of the oligonucleotide while leaving the central portion as native DNA or containing RNase H-compatible substitutions (57). The RNase H enzyme recognizes the ASO:RNA duplex and catalytically cleaves the RNA substrate.
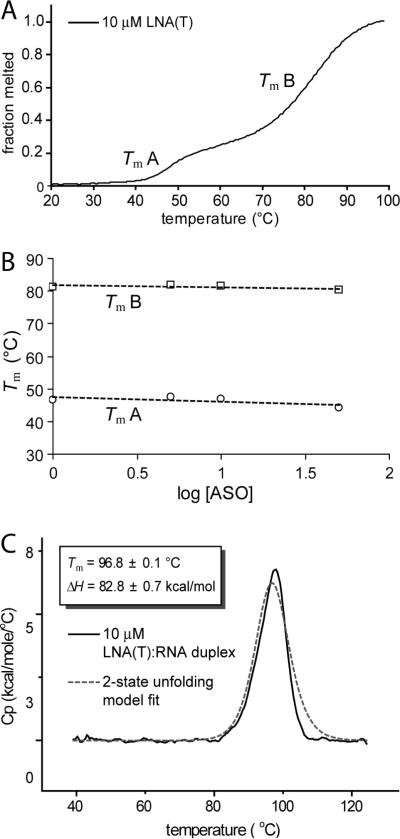
Oligonucleotides consisting of repeating GCT sequence are partially self-complementary and have potential to form hairpin structures (58, 59). Hairpin formation might affect recognition of the mRNA target. To investigate the stability of ASO self-structures and ASO:RNA duplexes, we used UV melt analysis and differential scanning calorimetry (DSC) (Figure S1). DSC was useful for measuring very high Tm values because it can scan samples under pressure at temperatures up to 130°C. The temperature corresponding to a maximum change in heat capacity (Cp) of the ASO:RNA duplex can be taken as the Tm (60, 61).
Selective inhibition of mutant HTT expression with LNA-modified ASOs
LNAs were evaluated in patient-derived GM04281 fibroblasts (Figures 2, S2 and S3). GM04281 cells harbor a normal HTT allele of 17 CAG repeats and a mutant allele with 69 CAG repeats and we have used this cell line previously to investigate allele-selective inhibition of HTT (33–35).
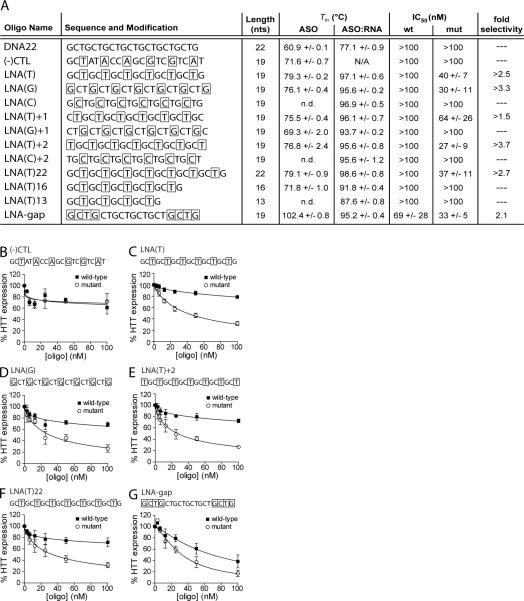
Figure 2. Selective inhibition of mutant HTT expression is observed with LNA-modified ASOs of varying sequence, length or modification configuration.
(A) Properties of LNA-modified ASOs that are complementary to the HTT mRNA CAG repeat sequence. ASO, antisense oligonucleotide alone; ASO:RNA, antisense oligonucleotide duplexed with complementary RNA. DNA22 is an unmodified DNA oligo and (−)CTL is a scrambled sequence containing LNA at every third base. Bases enclosed by boxes indicate LNA modification position. Tm values were experimentally determined by UV melt analysis for ASOs and differential scanning calorimetry (DSC) for ASO:RNA duplexes. Error is standard deviation. IC50 values are calculated from Western blot quantification. Values are reported in mean +/− standard error of the mean (SEM). N/A, not applicable; n.d., not determined due to insufficient cooperativity. (B–G) HTT protein expression in patient-derived fibroblast cells containing 69 CAG repeats in the mutant allele (GM04281) in response to increasing doses of control and inhibitory ASOs. Quantification and a non-linear fit of HTT expression levels from multiple dose responses are plotted. Sequence and modification configuration are indicated above the graph. Error bars are SEM.
We tested a variety of LNAs that differed in length, position of LNA substitution, and register of the sequence relative to the target triplet repeat. Four ASOs, LNA(T), LNA(G), LNA(T)+2 and LNA(T)22, inhibited HTT expression in an allele-selective fashion. Quantification of Western blots after dose response treatments revealed potencies ranging from IC50 values of 27 to 40 nM and selectivities of inhibition for mutant versus wild-type HTT by as much as > 3.7 fold (Figure 2A).
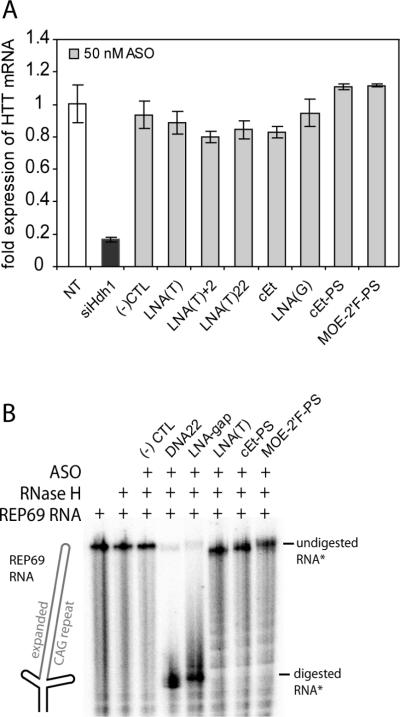
LNA-gap, which contains a central DNA portion capable of recruiting RNase H, inhibited HTT expression with lower selectivity (2.1 fold). We noted, however, that the introduction of LNA-gap into cells caused a moderate amount cell death (Figure S4). One explanation is that the ability to recruit RNase H might cause greater inhibition of other genes that contain CAG repeats since gapmer designs would cause catalytic degradation of RNA targets. Over 300 exonic sequences in humans are predicted to contain six or more repeated CAG units (62). Inhibition of both HTT alleles is not the cause of toxicity since siHdh1, an siRNA inhibiting both alleles and directed against sequences downstream of the repeat region (63) (see Figure 6A), showed no toxicity.
Figure 6. Allele-selective ASOs leave HTT mRNA intact and do not support RNase H activity in vitro.
(A) HTT mRNA levels are not substantially affected by treatment with allele-selective ASOs as determined by quantitative RT-PCR. Amplification of HTT cDNA was normalized to GAPDH amplification. NT, non-transfected. siHdh1 (transfected at 25 nM) is a duplex siRNA targeted to sequence downstream of the HTT CAG repeat (63). Error bars are standard deviation of triplicate experiments. (B) Allele-selective ASOs do not support RNase H cleavage in vitro. Allele-selective ASOs or control oligonucleotides were incubated with an in vitro transcribed and 5′-radiolabeled 69 CAG repeat-containing HTT mRNA 5′ end transcripts followed by treatment with RNase H enzyme. Reaction products were resolved on a denaturing polyacrylamide gel then visualized by phosphorimager.
Although only four LNA-modified ASOs exhibited clear allele-selective inhibition, all 19 and 22 nucleotide LNAs hybridized to CAG repeat RNA with comparably high Tm values. These data indicate that the potential for strong hybridization may be necessary but not sufficient for allele-selectivity (Figure 2A). Loss of inhibition by shortening of the benchmark ASO [LNA(T)16 or LNA(T)13] correlated with reduced Tm values for hybridization to a complementary RNA. Although substitution of the T or G bases resulted in inhibitory and allele-selective ASOs, modification of cytosine positions with LNA did not yield inhibitory oligonucleotides. Interestingly, these same two oligonucleotides, LNA(C) and LNA(C)+2, were observed to have poor cooperative melting and low hyperchromicity, which prevented accurate Tm determination. Poor cooperativity during thermal denaturation is indicative of low secondary structure or a population of conformationally irregular, non-homogeneous structures, such as aggregates (64, 65), suggesting a role for the degree and nature of self-interactions in effective inhibition with LNA-modified ASOs that target CAG repeats.
Effect of other bridged nucleic acid (BNA) and 2′ sugar modifications on allele-selective inhibition
To broaden the base of compounds available for development, obtain structure activity relationships for allele selective inhibition, and gain additional mechanistic insights, we tested oligomers containing other chemically modified nucleotides. We evaluated inhibition of HTT expression with oligonucleotides containing ENA, cET, carba-LNA, 2′F-RNA, 2′F-ANA, or MOE modifications (Figures 3, S2 and S5).
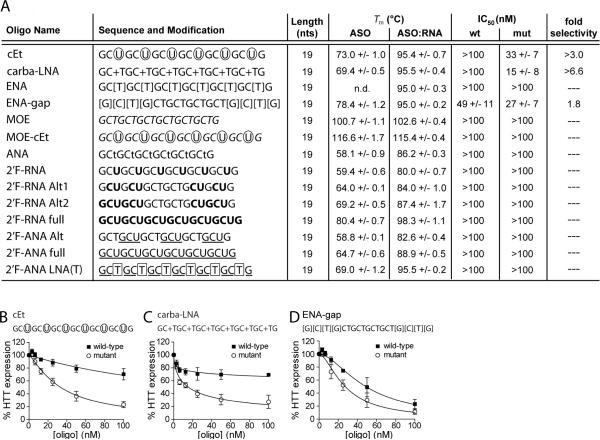
Figure 3. The BNA modifications LNA, cEt, and carba-LNA support allele-selective HTT inhibition.
(A) Properties of ASOs containing various modifications. ASO, antisense oligonucleotide alone; ASO:RNA, antisense oligonucleotide duplexed with complementary RNA. Position of nucleotide modifications within the ASO sequence are indicated as follows: boxed, LNA; circled, cEt; preceded by a “+”, carba-LNA; bracketed, ENA; italicized, MOE; lowercase, ANA; bold, 2'F-RNA; underlined, 2'F-ANA. Tm values were experimentally determined by UV melt analysis for ASOs and differential scanning calorimetry (DSC) for ASO:RNA duplexes and the MOE and MOE-cEt ASOs. Error is standard deviation. IC50 values are calculated from Western blot quantification. Values are reported in mean +/− standard error of the mean (SEM). n.d., not determined due to insufficient cooperativity. (B–D) HTT protein expression in patient-derived fibroblast cells containing 69 CAG repeats in the mutant allele (GM04281) in response to increasing doses of inhibitory modified ASO. Quantification and a non-linear fit of HTT expression levels from multiple dose responses are plotted. Sequence and modification configuration are indicated above the graph. Error bars are SEM.
Quantification of Western blots after dose response treatment with cET and carba-LNA revealed >3.0 and >6.6 fold selectivity, respectively (Figure 3). The IC50 for carba-LNA was 15 nM while the value for cET was 33 nM. The ENA-gap ASO inhibited expression of both alleles with poor selectivity and moderate toxicity (Figure S4), a result similar to that achieved using the LNA-modified gapmer LNA-gap (Figure 2). The remaining modified ASOs did not yield substantial inhibition of either wild-type or mutant HTT expression (Figure S5).
It is interesting that some modifications yielded potent allele-selective inhibitors whereas others did not. Several of the ASOs showed reduced hybridization affinities for complementary RNA compared to BNA-modified oligonucleotides (Figure 2A and 3A), especially the ANA, 2′F-RNA, 2′F-ANA, and 2′-FANA full designs. Reduced affinity for the target sequence provides one simple explanation for lack of inhibition.
Other inactive ASOs, however, such as fully modified 2′F-RNA full and 2′F-ANA LNA(T), had Tm values that were similar to active ASOs. For example, inactive 2′F-ANA LNA(T) had a melting temperature comparable to LNA(T) but was not inhibitory. The 2′F-ANA modification is known to acquire a south/east pucker and prefer a B-form helix (66) and thus acquires more DNA-like properties but with less flexibility (67). These properties may disrupt a unique helical structure that is induced by periodically spaced BNA-modified oligonucleotides and required for inhibition of HTT expression (68). Indeed, LNA influences adjacent nucleotides by inducing them into a northern conformation (42).
For the ENA ASO, which also showed poor cooperativity in UV melt analysis, flexibility and self-structure may play a role in its lack of inhibition. The modification is considered intrinsically more flexible due to a longer bridge and a greater rotation around its δ bond angle (48). The 2′-O-(2-methoxyethyl) modification (MOE) combined with cEt (MOE-cEt) resulted in similar Tm values for the ASO alone or in combination with RNA (Figure 3A), suggesting that oligonucleotide self-structure may be too stable and thus reduce productive hybridization with complementary RNA in the cell.
Another possible contribution to the differences observed between modifications is that, although all modified oligomers contain similar phosphodiester backbones, the ribose modifications may lead to different levels of cellular uptake when transfected with different transfection reagents. To investigate this possibility, we tested delivery of duplex RNA siHdh1 and the LNA(T), MOE, 2'-F-RNA full and 2'-F-ANA full ASOs using the delivery agents Lipofectamine RNAiMAX (Invitrogen), TriFECTin (Integrated DNA Technologies, Inc.), TransIT-Oligo (Mirus), Oligofectamine (Invitrogen), and PepMute (SignaGen) following the manufacturer's recommended protocols (Supplemental Figure S6).
We observed that several lipids yielded poor inhibition of HTT expression by LNA(T), demonstrating that the choice of lipid does affect whether activity is observed. By contrast, use of the peptide-based delivery agent PepMute not only yielded efficient inhibition of HTT expression by LNA(T), but also showed that inhibition by MOE was also achievable. These data make the important general point that choice of transfection reagent can affect comparisons between oligomer chemistries. Results should be interpreted conservatively. Positive data indicates that inhibition of gene expression can be achieved and encourages further experimentation, but negative data can not always be assumed to indicate that an oligomer cannot function under any experimental conditions.
Effect of phosphorothioate substitution
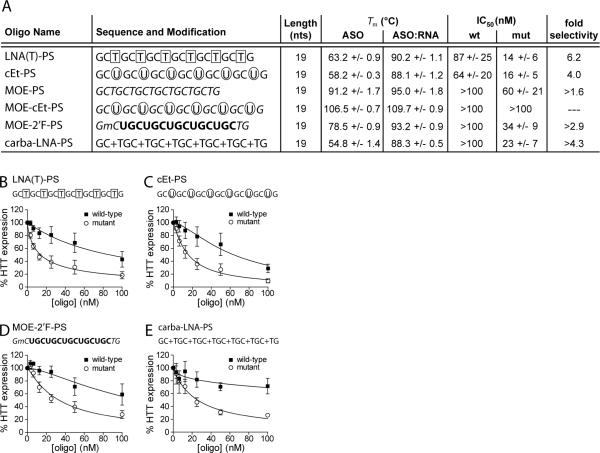
Phosphorothioate (PS) modifications confer favorable pharmacological properties upon ASOs including increased stability against nuclease digestion and improved biodistribution (38). To determine whether introduction of PS linkages was compatible with allele-selective inhibition, we replaced all phosphodiester bonds in several oligonucleotides with the PS modification (Figure 4). Three of the ASOs, LNA(T)-PS, cEt-PS and carba-LNA-PS, were based upon phosphodiester designs already shown to be allele-selective (Figures 2 and 3). One design was novel, MOE-2'F-PS, and combined two modifications, 2′FRNA and MOE, not previously observed to yield allele-selective ASOs alone (Figure 3).
Figure 4. Phosphorothioate backbone modification is compatible with allele-selective inhibition.
(A) Properties of CTG-repeat ASOs containing select modifications and a uniform phosphorothioate (PS) backbone. ASO, antisense oligonucleotide alone; ASO:RNA, antisense oligonucleotide duplexed with complementary RNA. Modification positions within the sequence are indicated as follows: boxed, LNA; circled, cET; italicized, MOE; bold, 2'F-RNA; preceded by a “+”, carba-LNA; mC, 5-methyl cytosine. Tm values were experimentally determined by UV melt analysis for ASOs and differential scanning calorimetry (DSC) for ASO:RNA duplexes and the MOE-PS and MOE-cEt-PS ASOs. Error is standard deviation. IC50 values are calculated from Western blot quantification. Values are reported in mean +/− standard error of the mean (SEM). (B–E) HTT protein expression in patient-derived fibroblast cells containing 69 CAG repeats in the mutant allele (GM04281) in response to increasing doses of inhibitory phosphorothioate-modified ASOs. Quantification and a non-linear fit of HTT expression levels from multiple dose responses are plotted. Sequence and modification configuration are indicated above the graph. Error bars are SEM.
PS-substituted ASOs LNA(T)-PS, cEt-PS, MOE-2'F-PS, and carba-LNA-PS achieved potent and selective inhibition of HTT (Figure 4A). This inhibition was observed even though the four ASOs possessed reduced melting temperatures compared to analogous active ASOs that have phosphodiester linkages. MOE-PS and MOE-cEt-PS showed little selectivity, although they displayed mild inhibition (Figure S7). MOE-PS and MOE-cEt-PS had similar melting temperatures for the ASO alone and the corresponding ASO:RNA duplex, suggesting highly stable self-structure. All phosphorothioate ASOs displayed some toxicity (Figure S4). Although PS-modified oligonucleotides often show some level of toxicity in tissue culture cells, they are generally non-toxic when introduced into live animals at effective doses (69–71). These data demonstrate that allele-selective inhibition is compatible with the pharmacologically favorable PS modification.
Selective inhibition in cell lines with minimal CAG repeats
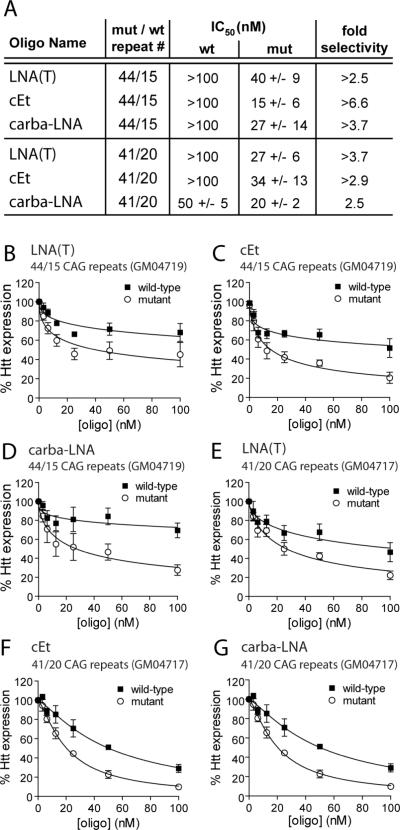
The mean CAG repeat length in HD patients is 44 (72). Patient-derived fibroblasts containing 69 CAG repeats in the mutant allele and 17 in the normal allele (GM04281) were employed for initial studies because separation of the two proteins is relatively straightforward, facilitating the screening of numerous compounds. To determine whether ASOs would also selectively inhibit mutant HTT expression when CAG repeat length was more characteristic of most HD patients, we tested inhibition by the most promising compounds in GM04719 (44 mutant repeats/15 wild-type repeats) or GM04717 (41 mutant repeats/20 wild-type repeats) patient-derived fibroblasts.
LNA(T), cET, and carba-LNA were tested in both the 44 repeat and 41 repeat cell lines and achieved allele-selective inhibition of HTT (Figure 5A–G). IC50 values for inhibition of mutant HTT ranged from 15 to 40 nM (Figure 5A–G and Figure S8). The allele-selectivity of inhibition varied between 2.5 fold to >6.6-fold. These data demonstrate that selectivity can be achieved in cell lines with mutant HTT alleles containing relatively few repeats.
Figure 5. Allele-selective ASOs discriminate between mutant and wild-type alleles in patient-derived fibroblasts with 44 or 41 CAG repeats in the mutant HTT mRNA.
(A) Allele-selectivity and potency of three selective ASOs when targeting mutant HTT mRNAs with only 44 or 41 CAG repeats. IC50 values are calculated from Western blot quantification. Values are reported in mean +/− standard error of the mean (SEM). (B–G) HTT protein expression in patient-derived fibroblast cells containing 44 (GM04719) or 41 (GM04717) CAG repeats in the mutant allele in response to increasing doses of inhibitory ASOs. Quantification and a non-linear fit of HTT expression levels from multiple dose responses are plotted. Error bars are SEM.
Specificity for the expanded CAG repeat HTT allele
Other human genes also have tracts of CAG repeats, although these tracts are usually much shorter than those observed for mutant HTT mRNA (62). A major concern for using ASO's to selectively block mutant HTT expression would be off-target inhibition of other genes. We have previously shown that peptide-PNA conjugates and the benchmark ASO LNA(T) do not affect endogenous CAG-repeat gene expression (33). To evaluate the potential for off-target effects with allele-selective ASOs composed of various chemical modifications, we treated patient-derived GM04281 fibroblasts and monitored expression of repeat-containing genes. Western blot analysis showed no effect for these ASOs on the expression levels of TATA-box binding protein (TBP), which harbors CAG repeat tracts of up to 19 repeats in the coding region, or forkhead box protein p2 (FOXP2), which contains a mixed stretch of 40 CAG and CAA repeats in the 5′-UTR (Figure S9). Ultimately, the possibility of toxicity due to off-target effects must be tested in vivo, but our experiments suggest that there is a window for achieving specific inhibition of mutant HTT. Efficient inhibition by ASOs may require CAG repeat expansions beyond a certain threshold length and perhaps also be dependent on the context of the surrounding sequence.
The effect of allele-selective ASOs on HTT mRNA levels and RNase H activity
Antisense oligonucleotides targeted to mRNA typically inhibit gene expression by inducing degradation of or by blocking translation (38, 57, 69). To investigate the mechanism of allele-selective ASOs we examined their effect on levels of HTT mRNA (Figure 6A). As a positive control, we demonstrated that transfection with 25 nM of siHdh1, a duplex RNA complementary to HTT mRNA at a sequence downstream of the CAG repeat (63), reduced total HTT mRNA levels to <20%. Treatment with a scrambled LNA control, (−) CTL, and a variety of allele-selective ASOs at 50 nM had little affect on HTT mRNA levels. Thus, RNA degradation does not appear to be a major determinant of allele-selective HTT inhibition.
We also tested the ability of allele-selective ASOs to support RNase H activity in vitro (Figure 6B). The 5′-end of mutant HTT mRNA containing 69 CAG repeats flanked by non-repetitive sequences was cloned as cDNA from GM04281 cells and in vitro transcribed (Figure S10). The resultant REP69 RNA was 5′-radiolabeled, hybridized to ASO, and treated with RNase H. The non-complementary (−)CTL oligonucleotide had no effect on RNA stability. As expected the DNA22 and LNA-gap yielded efficient cleavage. Cleavage of the RNA substrate was not readily detectable when RNase H was added in combination with the allele-selective LNA(T), cEt-PS and MOE-2'F-PS oligonucleotides. These results are consistent with the conclusion that allele-selective ASOs targeting the expanded HTT CAG repeat may function through a steric translational block mechanism rather than RNase H activity.
Properties of an allele-selective ASO
Allele-selective ASOs exhibit high melting temperatures both alone and when hybridized with complementary RNA (Figures 1A, 2A and 3A). To better characterize the properties of allele-selective ASOs, we monitored melting of the benchmark antisense oligonucleotide LNA(T) either alone or annealed to fully complementary RNA. In the absence of its RNA partner, LNA(T) alone melted with two clear transitions (Figure 7A). Tm values for each transition were not dependent on concentration of LNA(T) (Figure 7B), indicating that the observed transitions were due to intramolecular structure formation. These intramolecular structures are most likely hairpins. Thermal denaturation of the LNA(T):RNA duplex using DSC revealed a very stable complex with a single unfolding transition and a Tm of ~97°C (Figure 7C). These observations point to the conclusion that LNA(T) can form stable intramolecular interactions, but that the interactions do not interfere with the formation of a stable duplex with complementary RNA.
Figure 7. The allele-selective ASO LNA(T) forms a stable self-structure and RNA:ASO duplex.
(A) UV melt analysis of LNA(T), an allele-selective inhibitor of HTT expression, in the absence of an RNA complement. (B) UV melts of LNA(T) at concentrations spanning almost two orders of magnitude show no concentration dependence for Tm, suggesting an intramolecular hairpin folded structure. Tm A and Tm B correspond to the indicated transitions in panel A. (C) Differential scanning calorimetry (DSC) of LNA(T) base-paired to a fully complementary RNA. The ASO:RNA duplex is exceptionally stable, with a Tm of ~ 97°C. Values were fit to a 2-state unfolding model to calculate ΔH at the Tm.
Cumulative binding of multiple ASOs as a mechanism for allele-selectivity
Mutant HTT repeats have more sites for ASO binding than do wild-type repeats. For example, a 69 CAG repeat sequence can bind up to ten 20 base ASOs, whereas a 17 base wild-type repeat sequence can bind no more than two to three. Therefore, one simple mechanism that might contribute to selective inhibition would be binding of more ASOs to the mutant repeat than to the wild-type.
We performed in vitro binding assays to gain insights into the varying levels of allele-selectivity elicited by ASOs (Figure 8). Complementary DNA (cDNA) of mRNA 5′-ends from mutant and normal HTT mRNA were cloned from GM04281 cells and in vitro transcribed. Both RNAs begin at the start codon and continue past the CAG repeat. The cloned region was designed to incorporate flanking RNA sequence to preserve any surrounding mRNA structure that might be critical to native folding of the repeat (Figure S10).
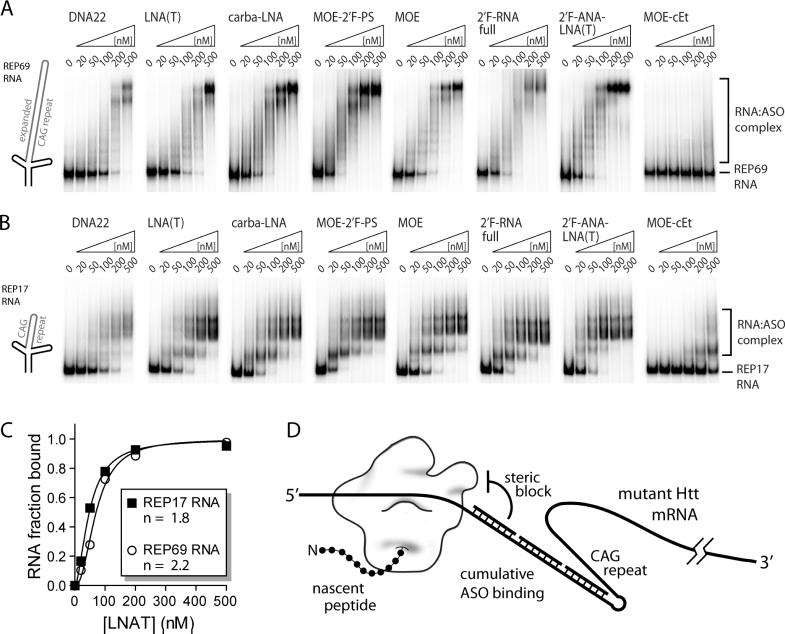
Figure 8. Multiple ASOs bind CAG repeat-containing HTT 5′-end transcripts in vitro.
(A–B) Electrophoretic mobility shift assays (EMSA) demonstrate cooperative and near stoichiometric binding of multiple ASOs to CAG repeat sequence in HTT mRNA 5′ end transcripts, as well as a case where no binding is observed. In vitro transcribed and 5′-radiolabeled wild-type and mutant 5′-end HTT mRNA transcripts were incubated with increasing concentrations of ASO. RNA:ASO complexes were resolved on native polyacrylamide gels and visualized by phosphorimager. ASO concentrations are indicated above the gels and shifted bands identified to the right. (C) Quantification of LNA(T) binding REP17 and REP69 HTT mRNA transcripts indicates cooperative ASO binding. ASO-bound RNA in gel shifts from panels A and B were quantified and plotted as a function of concentration. Fitting to the Hill equation revealed a sigmoidal binding curve and Hill coefficients (n) near 2, suggesting cooperative ASO binding. (D) Putative steric translational blocking mechanism. Translation is repressed on a mutant expanded CAG repeat HTT mRNA due to a proposed steric inhibition of translation by cumulative ASO binding.
Mutant RNA (REP69) and normal RNA (REP17) were 5′-radiolabeled, refolded, and mixed with increasing concentrations of ASO followed by resolution of ASO:RNA complexes on native polyacrylamide gels. The different ASOs appeared to bind with varying affinities. Interestingly, the MOE-cEt ASO did not bind REP69 and only poorly bound REP17 RNA. Since this oligonucleotide exhibited such stable self-structure (Figure 3A), it may be unable to overcome kinetic or thermodynamic barriers to bind the target RNA.
Binding by multiple oligomers could be observed for most ASOs. Discrete bands corresponding to individual binding events was observed over the course of the titration. This resulted in 10 or more ASOs hybridizing to the REP69 RNA and between 2–3 ASOs for the REP17 RNA, both near upper limits of binding events predicted considering ASO and target CAG repeat lengths. These findings suggest that the quantity of ASOs able to bind a folded HTT mRNA in vitro are only limited by the number of CAG repeats available, consistent with the CAG repeat being a semi-stable but accessible hairpin structure (58, 59, 73). Quantification of gels followed by fitting to a Hill equation revealed strong sigmoidal curves and Hill coefficients around 2, indicative of cooperative ASO binding (Figure 8C and S11). While the situation inside cells is not known, these data from in vitro assays suggest that cumulative and cooperative binding of ASOs to longer mutant CAG repeats may contribute to increased recognition and allele-selectivity (Figure 8D).
DISCUSSION
Development of safe and effective strategies for reducing expression of HTT is an important goal for HD research. siRNAs or ASOs offer nucleic-acid based strategies for achieving this goal. While nucleic acids are not a traditional approach towards development of therapies for brain disorders, recent advances in their chemistry, pharmacology, and delivery have made them a realistic option for diseases like HD where patients have no curative treatments available (74, 75).
It is possible that useful siRNAs or ASOs will be effective even if they cause equal reduction of both HTT alleles (25). However, given the potential unknown side-effects of long-term inhibition of normal HTT expression in patients with HD, allele-selective strategies offer a useful addition to the treatment options being considered (26). Allele-selective siRNAs that exploit single nucleotide polymorphisms for specificity are the subject of intense interest by several laboratories (28, 30–32).
While siRNAs and antisense oligonucleotides inhibit gene expression through similar mechanisms, they have different strengths. siRNAs benefit from an endogenous gene regulatory pathway, RNA interference (RNAi), and are incorporated into the RNA-induced silencing complex (RISC) to guide degradation of complementary RNA. These properties can make siRNAs robust inhibitors of gene expression and help protect them from degradation (76).
ASOs offer an alternate approach. Because they are single-stranded they can be less expensive to manufacture. They do not need to be incorporated into a protein complex, possibly leading to more predictable off-target effects and the potential for more extensive chemical modification (69). ASOs have been thoroughly tested in the clinic and may prove to be a more direct route to a nucleic acid-based therapeutic for allele-specific inhibition of mutant HTT expression (26, 37–41). ASOs, by virtue of being half the size of duplex RNAs, may show better distribution in vivo, although this possibility has not yet been demonstrated. Pursuing both the ASO and siRNA strategies should maximize the likelihood that a useful agent will eventually benefit HD patients.
We have identified ten chemically-modified ASOs that selectively inhibited expression of the mutant HTT allele. A number of chemical modifications to the ribose were compatible with allele-selective inhibition, including LNA, cEt, carba-LNA, MOE, and 2'F-RNA, and phosphodiester internucleotide linkages could be substituted with phosphorothioate. When three ASOs were tested in patient-derived cell lines containing fewer CAG repeats that are more representative of the HD patient population, they all showed allele-selective inhibition. At least five ASOs of diverse chemistry appeared specific for the expanded CAG repeat of HTT since they had no effect on two unrelated genes containing substantial CAG repeat tracts. Although tentative, design rules for allele-selective ASOs targeting expanded CAG repeats should include high hybridization affinity tempered with moderate intramolecular self-structure. Among the allele-selective ASOs, bridged nucleic acids were highly successful and should be considered as lead modifications.
It is not always straightforward to understand why some ASOs are more effective than others, especially with such a wide variety of modification types and configurations under evaluation. A number of factors can complicate an oligonucleotide's ability to reach its target RNA and inhibit gene expression. Simple explanations might include poor transfection or release from endosomes after cell entry. Future studies will be needed to more clearly determine why some chemical modifications were ineffective and why even among the bridged nucleic acid modifications there were varying degrees of inhibition and selectivity. Nonetheless, our data provide a broad choice of compounds for testing in vivo or for further optimization as drug candidates.
Our data suggest that allele-selective inhibition can be achieved by a mechanism involving a steric block of translation (Figure 8D). The most successful allele-selective ASOs did not have the central DNA bases found in gapmer antisense oligonucleotides and did not support RNase H activity in vitro. Allele-selective ASOs also had little or no effect on HTT mRNA levels. We previously reported that a fully complementary CTG:CAG repeat small duplex RNA (siREP) targeted to the CAG repeat, which presumably induces RNA cleavage in the HTT repeat region via the RNA-induced silencing complex (RISC), was a robust inhibitor of HTT expression but very non-selective (33, 34). Taken together, these results suggest that directed cleavage of RNA does not contribute significantly to the mechanism of allele-selective inhibition.
In support of a translational inhibition model for allele-selectivity, our ASOs bound CAG repeat RNA in a cooperative manner in vitro, with multiple ASOs assembling onto all of the potentially available CAG repeat sequence. Thus, selective inhibition could arise from greater accumulation of ASOs on the expanded CAG repeat HTT mRNA and preferentially block translation of the mutant HTT protein. Cooperative binding could also contribute to the specificity of allele-selective ASOs for the HTT mRNA by enhancing affinity with each binding event as the RNA structure is invaded and becomes increasingly more accessible. Investigations of the CAG repeat structure for other mRNAs suggests that HTT mRNA may well have unique properties that help facilitate cooperative ASO binding (59, 73). In addition, effective translational repression by ASOs is often achieved when targeting near the start codon of an mRNA (45, 77). The expanded CAG repeat of HTT is near the 5′-end of the mRNA with only 51 nucleotides between the translational start site and the beginning of the CAG repeat tract, making it a good candidate for translational repression by antisense oligonucleotides.
Our findings suggest that targeting repeats may serve as a general strategy for developing nucleic acid-based therapeutics for disease-related genes that harbor simple repeat expansions. A recent report demonstrated the therapeutic potential of a CAG-repeat morpholino targeted to DM protein kinase (DMPK) mRNA, which contains a CUG repeat expansion in myotonic dystrophy type 1 (DM1) (36). The morpholino disrupted ribonucleoprotein complexes formed between the expanded CUG repeat of DMPK mRNA and muscleblind-like 1 (MBNL-1) protein, an interaction directly implicated in disease. Competition with the complementary morpholino released MBNL-1 and rescued the myotonic dystrophy phenotype in a mouse model of DM. Most trinucleotide and some tetranucleotide repeat expansions form semi-stable hairpin structures, much like that observed in the CAG repeat region of HTT (58, 59, 73). Thus, simple repeat sequences should be generally amenable to targeting with complementary ASOs to selectively modulate gene expression and ultimately provide new hope for currently untreatable genetic diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jiaxin Hu and Masayuki Matsui for helpful discussions and Jonathan K. Watts for critical reading of the manuscript. We also thank members of the laboratory of Dr. Rama Ranganathan for help with differential scanning calorimetry experiments.
FUNDING This work was supported by the National Institutes of Health (GM 73042 to DRC and 1F32HD060377-01A1 to KTG) and the Robert A. Welch Foundation (I-1244 and I-1336).
ABBREVIATIONS
- HTT
huntingtin
- ASO
antisense oligonucleotide
- HD
Huntington's Disease
- SNP
single nucleotide polymorphism
- siRNA
small interfering RNA
- LNA
locked nucleic acid
- BNA
bridged nucleic acid
- DSC
differential scanning calorimetry
- UV
ultraviolet
- qPCR
quantitative polymerase chain reaction
- EMSA
electrophoretic mobility shift assay
Footnotes
SUPPORTING INFORMATION Derivation of experimental Tm values from UV melt analysis and DSC (Figure S1), representative Western blots for quantification of HTT inhibition with allele-selective or control ASOs (Figure S2) or ASOs that showed poor inhibition or selectivity (Figure S3, S5 and S7) in patient derived cells with 69 CAG repeats in the mutant allele, viability of patient-derived fibroblasts after treatment with various ASOs (Figure S4), inhibition of HTT expression with several ASOs using different transfection reagents (Figure S6), representative Western blots for quantification of HTT inhibition in patient-derived cells with 44 or 41 CAG repeats (Figure S8), Western blots showing specificity of allele-selective ASOs for the mutant HTT gene (Figure S9), sequence and predicted secondary structure of the 5′-end of HTT mRNA transcripts (Figure S10), and quantification of multiple ASOs that appear to cooperatively bind REP17 and REP69 HTT RNA transcripts (Figure S11). This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Ann. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Walker FO. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald ME, Ambrose CM, Duyao MP, Myers RH, Lin C, Srinidhi L, Barnes G, Taylor SA, James M, Groot N, MacFarlane H, et al. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 4.Rubinsztein DC. Molecular biology of Huntington's disease (HD) and HD-like disorders. In: Pulst S, editor. Genetics of Movement Disorders. Academic Press; California: 2003. pp. 365–377. [Google Scholar]
- 5.Rubinsztein DC, Leggo J, Coles R, Almqvist E, Biancalana V, Cassiman JJ, Chotai K, Connarty M, Crauford D, Curtis A, et al. Phenotypic characterization of individuals with 30–40 CAG repeats in the Huntington disease (HD) gene reveals HD cases with 36 repeats and apparently normal elderly individuals with 36–39 repeats. Am. J. Hum. Genet. 1996;59:16–22. [PMC free article] [PubMed] [Google Scholar]
- 6.McNeil SM, Novelletto A, Srinidhi J, Barnes G, Kornbluth I, Altherr MR, Wasmuth JJ, Gusella JF, MacDonald ME, Myers RH. Reduced penetrance of the Huntington's disease mutation. Hum. Mol. Genet. 1997;6:775–779. doi: 10.1093/hmg/6.5.775. [DOI] [PubMed] [Google Scholar]
- 7.Kremer B, Goldberg P, Andrew SE, Theilmann J, Telenius H, Zeisler J, Squitieri F, Lin B, Bassett A, Almqvist E, Bird TD, Hayden MR. A worldwide study of the Huntington's disease mutation. The sensitivity and specificity of measuring CAG repeats. N. Engl. J. Med. 1994;330:1401–1406. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- 8.Snell RG, MacMillan JC, Cheadle JP, Fenton I, Lazarou LP, Davies P, MacDonald ME, Gusella JF, Harper PS, Shaw DJ. Relationship between trinucleotide repeat expansion and phenotypic variation in Huntington's disease. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 9.Rangone H, Humbert S, Saudou F. Huntington's disease: how does huntingtin, an anti-apoptotic protein, become toxic? Pathol. Biol. 2004;52:338–342. doi: 10.1016/j.patbio.2003.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Luo GR, Le WD. Collective roles of molecular chaperones in protein degradation pathways associated with neurodegenerative diseases. Curr. Pharm. Biotechnol. 2010;11:180–187. doi: 10.2174/138920110790909740. [DOI] [PubMed] [Google Scholar]
- 11.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 12.Hayden MR, Leavitt BR, Yasothan U, Kirkpatrick P. Tetrabenazine. Nat. Rev. Drug Disc. 2009;8:17–18. doi: 10.1038/nrd2784. [DOI] [PubMed] [Google Scholar]
- 13.Stewart JT. Treatment of Huntington's disease with clonazepam. South Med. J. 1988;81:102. doi: 10.1097/00007611-198801000-00029. [DOI] [PubMed] [Google Scholar]
- 14.Grote HE, Bull ND, Howard ML, van Dellen A, Blakemore C, Bartlett PF, Hannan AJ. Cognitive disorders and neurogenesis deficits in Huntington's disease mice are rescued by fluoxetine. Eur. J. Neurosci. 2005;22:2081–2088. doi: 10.1111/j.1460-9568.2005.04365.x. [DOI] [PubMed] [Google Scholar]
- 15.Peng Q, Masuda N, Jiang M, Li Q, Zhao M, Ross CA, Duan W. The antidepressant sertraline improves the phenotype, promotes neurogenesis and increases BDNF levels in the R6/2 Huntington's disease mouse model. Exp. Neurol. 2008;210:154–163. doi: 10.1016/j.expneurol.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Bae BI, Xu H, Igarashi S, Fujimuro M, Agrawal N, Taya Y, Hayward SD, Moran TH, Montell C, Ross CA, et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron. 2005;47:29–41. doi: 10.1016/j.neuron.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 18.DiProspero NA, Chen EY, Charles V, Plomann M, Kordower JH, Tagle DA. Early changes in Huntington's disease patient brains involve alterations in cytoskeletal and synaptic elements. J. Neurocytol. 2004;33:517–533. doi: 10.1007/s11068-004-0514-8. [DOI] [PubMed] [Google Scholar]
- 19.Charrin BC, Saudou F, Humbert S. Axonal transport failure in neurodegenerative disorders: the case of Huntington's disease. Pathol. Biol. 2005;53:189–192. doi: 10.1016/j.patbio.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 21.Gagnon KT. IDrugs; HD Therapeutics - CHDI Fifth Annual Conference; 2010. pp. 219–223. [PubMed] [Google Scholar]
- 22.Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 23.White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL, MacDonald ME. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 1997;17:404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- 24.Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM, Ge P, Vonsattel JP, Gusella JF, Joyner AL, et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 25.Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S, Bonvento G, Brouillet E, Luthi-Carter R, Hantraye P, Déglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 26.Scholefield J, Wood MJ. Therapeutic gene silencing strategies for polyglutamine disorders. Trends Genet. 2010;26:29–38. doi: 10.1016/j.tig.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc. Natl. Acad. Sci. USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Bilsen PH, Jaspers L, Lombardi MS, Odekerken JC, Burright EN, Kaemmerer WF. Identification and allele-specific silencing of the mutant huntingtin allele in Huntington's disease patient-derived fibroblasts. Hum. Gene Ther. 2008;19:710–718. doi: 10.1089/hum.2007.116. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Lebron E, Paulson HL. Allele-specific RNA interference for neurological disease. Gene Ther. 2006;13:576–581. doi: 10.1038/sj.gt.3302702. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Engelman J, Friedlander RM. Allele-specific silencing of mutant Huntington's disease gene. J. Neurochem. 2009;108:82–90. doi: 10.1111/j.1471-4159.2008.05734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E, Donato SD, Kaemmerer WF. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp. Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Matsui M, Corey DR. Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann. N.Y. Acad. Sci. 2009;1175:24–31. doi: 10.1111/j.1749-6632.2009.04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu J, Dodd DW, Hudson RH, Corey DR. Cellular localization and allele-selective inhibition of mutant huntingtin protein by peptide nucleic acid oligomers containing the fluorescent nucleobase [bis-o-(aminoethoxy)phenyl]pyrrolocytosine. Bioorg. Med. Chem. Lett. 2009;19:6181–6184. doi: 10.1016/j.bmcl.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corey DR. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swayze EE, Bhat B. The Medicinal Chemistry of Oligonucleotides. In: Crooke ST, editor. Antisense Drug Technology: Principles, Strategies, and Applications. Second ed. CRC Press; Boca Raton, FL: 2008. pp. 143–182. [Google Scholar]
- 39.Raal FJ, Santos RD, Blom DJ, Marais AD, Charng MJ, Cromwell WC, Lachmann RH, Gaudet D, Tan JL, Chasan-Taber S, et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: a randomised, double-blind, placebo-controlled trial. Lancet. 2010;375:998–1006. doi: 10.1016/S0140-6736(10)60284-X. [DOI] [PubMed] [Google Scholar]
- 40.Akdim F, Visser ME, Tribble DL, Baker BF, Stroes ES, Yu R, Flaim JD, Su J, Stein EA, Kastelein JJ. Effect of mipomersen, an apolipoprotein B synthesis inhibitor, on low-density lipoprotein cholesterol in patients with familial hypercholesterolemia. Am. J. Cardiol. 2010;105:1413–1419. doi: 10.1016/j.amjcard.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 41.Akdim F, Stroes ES, Sijbrands EJ, Tribble DL, Trip MD, Jukema JW, Flaim JD, Su J, Yu R, Baker BF, et al. Efficacy and safety of mipomersen, an antisense inhibitor of apolipoprotein B, in hypercholesterolemic subjects receiving stable statin therapy. J. Am. Coll. Cardiol. 2010;55:1611–1618. doi: 10.1016/j.jacc.2009.11.069. [DOI] [PubMed] [Google Scholar]
- 42.You Y, Moreira BG, Behlke MA, Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucl. Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obika S, Nanbu D, Hari Y, Andoh J, Morio K, Doi T, Imanishi T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 44.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 45.Braasch DA, Liu Y, Corey DR. Antisense inhibition of gene expression in cells by oligonucleotides incorporating locked nucleic acids: effect of mRNA target sequence and chimera design. Nucleic Acids Res. 2002;30:5160–5167. doi: 10.1093/nar/gkf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaur H, Arora A, Wengel J, Maiti S, Arora A, Wengel J, Maiti S. Thermodynamic, Counterion, and Hydration Effects for the Incorporation of Locked Nucleic Acid Nucleotides into DNA Duplexes. Biochemistry. 2006;45:7347–7355. doi: 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- 47.Morita K, Hasegawa C, Kaneko M, Tsutsumi S, Sone J, Ishikawa T, Imanishi T, Koizumi M. 2'-O,4'-C-ethylene-bridged nucleic acids (ENA): highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett. 2002;12:73–76. doi: 10.1016/s0960-894x(01)00683-7. [DOI] [PubMed] [Google Scholar]
- 48.Koizumi M, Morita K, Daigo M, Tsutsumu S, Abe K, Obika S, Imanishi T. Triplex formation with 2'-O,4'-C-ethylene-bridged nucleic acids (ENA) having C3'-endo conformation at physiological pH. Nucl. Acids Res. 2003;31:3267–3273. doi: 10.1093/nar/gkg416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, Witchell D, Swayze EE. Short antisense oligonucleotides with novel 2'–4' conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J Med Chem. 2009;52:10–13. doi: 10.1021/jm801294h. [DOI] [PubMed] [Google Scholar]
- 50.Prakash TP, Siwkowski A, Allerson CR, Migawa MT, Lee S, Gaus HJ, Black C, Seth PP, Swayze EE, Bhat B. Antisense oligonucleotides containing conformationally constrained 2',4'-(N-methoxy)aminomethylene and 2',4'-aminooxymethylene and 2'-O,4'-C-aminomethylene bridged nucleoside analogues show improved potency in animal models. J Med Chem. 2010;53:1636–1650. doi: 10.1021/jm9013295. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava P, Barman J, Pathmasiri W, Plashkevych O, Wenska M, Chattopadhyaya J. Five- and six-membered conformationally locked 2',4'-carbocyclic ribo-thymidines: synthesis, structure, and biochemical studies. J Am Chem Soc. 2007;129:8362–8379. doi: 10.1021/ja071106y. [DOI] [PubMed] [Google Scholar]
- 52.Allart B, Khan K, Helmut R, Schepers G, Hendrix C, Rothenbacher K, Seela F, Van Aerschot A, Herdewijn P. D-altritol nucleic acids (ANA): hybridisation properties, stability, and initital structural analysis. Chem. Eur. J. 1999;5:2424–2431. [Google Scholar]
- 53.Teplova M, Minasov G, Tereshko V, Inamati GB, Cook PD, Manoharan M, Egli M. Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA. Nat Struct Biol. 1999;6:535–539. doi: 10.1038/9304. [DOI] [PubMed] [Google Scholar]
- 54.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2'-deoxy-2'-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 55.Watts JK, Damha MJ. 2'F-Arabinonucleic acids (2'F-ANA) - History, properties, and new frontiers. Can. J. Chem. 2008;86:641–656. [Google Scholar]
- 56.Damha MJ, Wilds CJ, Noronha A, Brukner I, Borkow G, Arion D, Parniak MA. Hybrids of RNA and arabinonucleic acids (ANA and 2'F-ANA) are substrates of ribonuclease H. J. Am. Chem. Soc. 1998;120:12976–12977. [Google Scholar]
- 57.Bonham MA, Brown S, Boyd AL, Brown PH, Bruckenstein DA, Hanvey JC, Thomson SA, Pipe A, Hassman F, Bisi JE, et al. An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res. 1995;23:1197–1203. doi: 10.1093/nar/23.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Broda M, Kierzek E, Gdaniec Z, Kulinski T, Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–10882. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 59.Sobczak K, Michlewski G, de Mezer M, Kierzek E, Krol J, Olejniczak M, Kierzek R, Krzyzosiak WJ. Structural diversity of triplet repeat RNAs. J Biol Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chakrabarti MC, Schwarz FP. Thermal stability of PNA/DNA and DNA/DNA duplexes by differential scanning calorimetry. Nucl. Acids Res. 1999;27:4801–4806. doi: 10.1093/nar/27.24.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaur H, Wengel J, Maiti S. Thermodynamics of DNA-RNA heteroduplex formation: effects of locked nucleic acid nucleotides incorporated into the DNA strand. Biochemistry. 2008;47:1218–1227. doi: 10.1021/bi700996z. [DOI] [PubMed] [Google Scholar]
- 62.Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucl. Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Omi K, Hachiya NS, Tokunaga K, Kaneko K. siRNA-mediated inhibition of endogenous Huntington disease gene expression induces an aberrant configuration of the ER network in vitro. Biochem Biophys Res Commun. 2005;338:1229–1235. doi: 10.1016/j.bbrc.2005.10.061. [DOI] [PubMed] [Google Scholar]
- 64.Brion P, Michel F, Schroeder R, Westhof E. Analysis of the cooperative thermal unfolding of the td intron of bacteriophage T4. Nucl. Acids Res. 1999;27:2494–2502. doi: 10.1093/nar/27.12.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mergny J-L, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 66.Ikeda H, Fernandez R, Wilk A, Barchi JJ, Jr., Huang X, Marquez VE. The effect of two antipodal fluorine-induced sugar puckers on the conformation and stability of the Dickerson-Drew dodecamer duplex [d(CGCGAATTCGCG)]2. Nucleic Acids Res. 1998;26:2237–2244. doi: 10.1093/nar/26.9.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mangos MM, Min K-L, Viazovkina E, Galarneau A, Elzagheid MI, Parniak MA, Damha MJ. Efficient RNase H-directed cleavage of RNA promoted by antisense DNA or 2'F-ANA constructs containing acyclic nucleotide inserts. J. Am. Chem. Soc. 2002;125:654–661. doi: 10.1021/ja025557o. [DOI] [PubMed] [Google Scholar]
- 68.Kaur H, Wengel J, Maiti S. LNA-modified oligonucleotides effectively drive intramolecular-stable hairpin to intermolecular-duplex state. Biochem Biophys Res Commun. 2007;352:118–122. doi: 10.1016/j.bbrc.2006.10.155. [DOI] [PubMed] [Google Scholar]
- 69.Crooke ST. Progress in antisense technology. Annu. Rev. Med. 2004;55:61–95. doi: 10.1146/annurev.med.55.091902.104408. [DOI] [PubMed] [Google Scholar]
- 70.Henry SP, Templin MV, Gillett N, Rojko J, Levin AA. Correlation of Toxicity and Pharmacokinetic Properties of a Phosphorothioate Oligonucleotide Designed to Inhibit ICAM-1. Toxicol. Pathol. 1999;27:95–100. doi: 10.1177/019262339902700117. [DOI] [PubMed] [Google Scholar]
- 71.Ferdinandi ES, Vassilakos A, Lee Y, Lightfoot J, Fitsialos D, Wright JA, Young AH. Preclinical toxicity and toxicokinetics of GTI-2040, a phosphorothioate oligonucleotide targeting ribonucleotide reductase R2. Cancer Chemother. Pharmacol. 2010;66 doi: 10.1007/s00280-010-1473-z. advanced online. [DOI] [PubMed] [Google Scholar]
- 72.Andrew SE, Goldberg YP, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchman MA, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington's disease. Nat. Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 73.Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J Mol Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 74.Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010 doi: 10.1101/gad.1941310. in press. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G, Lobsinger CS, Ward CM, McAlonis-Downes M, Wei H, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 2006;116:2290–2296. doi: 10.1172/JCI25424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 77.Dias N, Dheur S, Nielsen PE, Gryaznov S, Van Aerschot A, Herdewijn P, Helene C, Saison-Behmoaras TE. Antisense PNA tridecamers targeted to the coding region of Ha-ras mRNA arrest polypeptide chain elongation. J Mol Biol. 1999;294:403–416. doi: 10.1006/jmbi.1999.3277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.