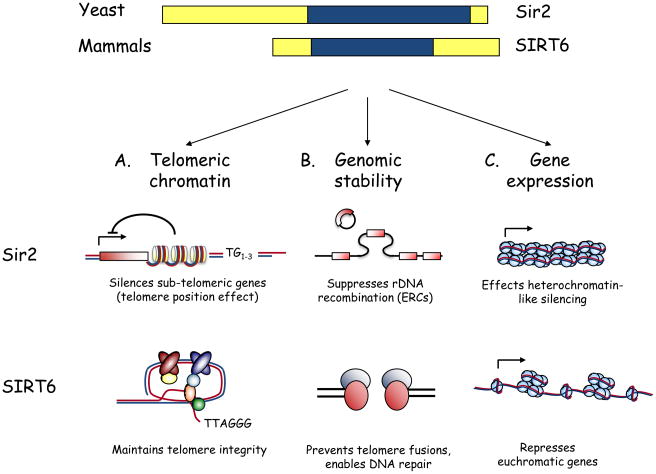
Figure 1.
From yeast to mammals: Parallel functions for Sir2 and SIRT6. A) Schematic of yeast Sir2 and mammalian SIRT6, highlighting the conserved central sirtuin domain (blue). B) Overlapping functions for Sir2 and SIRT6 in regulating telomeric chromatin, genomic stability, and gene expression. i. At yeast telomeres, histone deacetylation by Sir2 is required for telomere position effect, the reversible silencing of sub-telomeric gene expression. SIRT6 is required for generating a proper chromatin structure at human telomeres; this structure allows the binding of the telomere-processing factor WRN and prevents aberrant telomere metabolism. Whether SIRT6 is important for gene silencing due to telomere position effect in human cells is not currently known. ii. Yeast Sir2 maintains genomic stability by preventing recombination at the rDNA locus, thus suppressing formation of senescence-inducing extrachromosomal rDNA circles (ERCs). In mammalian cells, SIRT6 contributes to genomic stability by preventing chromosomal fusions between dysfunctional telomeres, by enabling efficient DNA repair, and perhaps by regulating H3K56 acetylation. iii. Sir2 is required for the transcriptional silencing of three heterchromatin-like domains in yeast: inactive mating-type loci, sub-telomeric DNA, and rDNA repeats. Although SIRT6 also represses gene expression, its reported effects thus far are at euchromatic regions of the genome (NF-κB and HIF1α target genes).