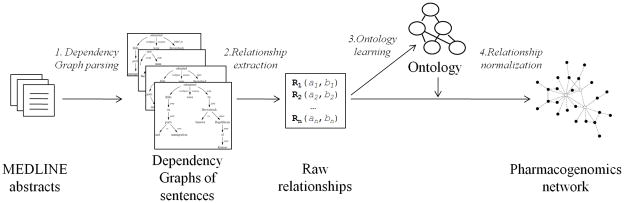
Figure 1.
Overview of our method to extract pharmacogenomics (PGx) relationships from text. The method has four steps. 1. We parse the text (Medline abstracts in this work) with the Stanford Parser to yield the Dependency Graph data structure that provides the syntactical structure of each sentence. 2. We identify PGx entities and their raw relationships—“raw” because their subject, object and type use natural language terms. 3. We processed these raw relationships to build (first run only) or refine (next runs) an ontology of PGx relationships. 4. For each of the raw relationships, we map them to the ontology and express them in normalized form. Normalized relationships create a network in which nodes are PGx entities and edges are relationships, both of which are associated with a precise semantics.