FIGURE 1.
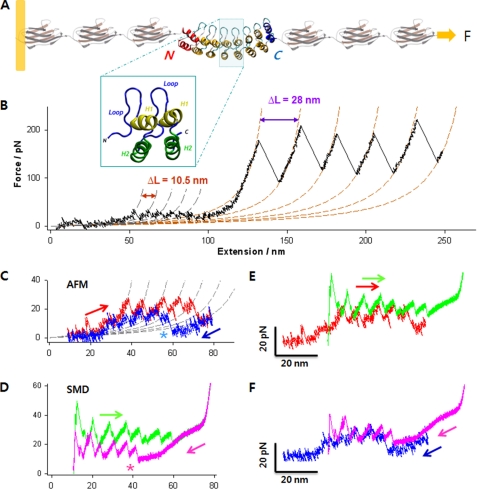
Complete mechanical unfolding and refolding of the NI6C-I27 chimeric protein. A, structure of the chimeric polyprotein designed for the AFM experiments. NI6C contains eight ARs (66). Two internal repeats are shown in the blue box, each composed of two α-helices (H1 and H2) and a loop. The full sequence is shown in supplemental Fig. S1D. B, representative unfolding trace of NI6C-I27 obtained at a stretching speed of 100 nm/s. The AFM data are fitted to two families of WLC curves, one with contour length increment ΔL = 10.5 nm and persistence length p = 0.78 nm (gray dashed lines) and the other with ΔL = 28 nm and p = 0.36 nm (orange dashed lines). These values of ΔL are consistent with the fully stretched lengths of one AR and one I27 domain, respectively; hence, the two families of peaks correspond to the unfolding of individual ARs of NI6C and of I27 domains, respectively. pN, piconewtons. C, measured unfolding (red) and refolding (blue) force extension traces of NI6C at 30 nm/s, fitted to a family of WLC curves (gray dashed lines) with ΔL = 10.5 nm and p = 0.86 nm. Note that following the complete unfolding of all ARs, the first refolding force peak appeared only after the molecule was partially relaxed (asterisk). D, simulated unfolding (green) and refolding (pink) force extension traces of NI6C. Similar to the AFM data, the first SMD refolding force peak occurred only after the molecule has been significantly relaxed (asterisk). E, comparison of the unfolding force extension traces by SMD (green) and AFM (red). The SMD trace was shifted to the right by 20 nm to compensate for the initial length of I27 modules that contribute to the extension in the AFM measurements but are absent in the SMD simulations. F, comparison of the refolding force extension traces by SMD (pink) and AFM (blue) following the unfolding of NI6C. The same 20-nm shift was applied to the SMD trace.