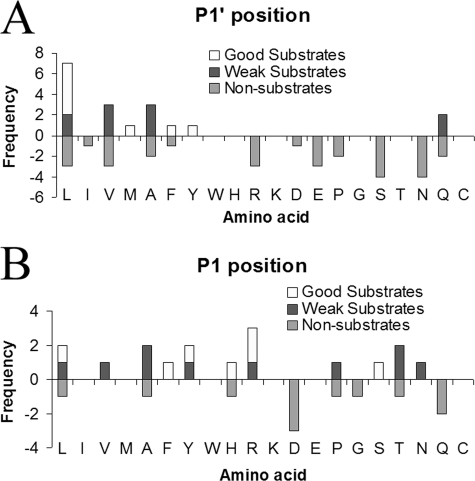
FIGURE 4.
Peptidomics analysis of CPA6 preferences at substrate P1′ and P1 positions. A, analysis of the P1′ residue. All good substrates have C-terminal Leu, Met, Phe, or Tyr, whereas weak substrates have C-terminal Leu, Val, Ala, or Gln. Many different C-terminal amino acids can be found in the non-substrate category. B, analysis of the P1 residue of peptides with permissive P1′ residues. Altogether, 28 peptides were detected with C-terminal hydrophobic residues (Leu, Ile, Val, Met, Ala, Phe, and Tyr) or Gln; all of these residues were found to be cleaved either for some peptides in the peptidomics analysis, or with small synthetic substrates. Of these 28, only 7 were good substrates, 10 were weak substrates, and 11 were not cleaved. Analysis of the sequences of these peptides indicates that most good substrates contain hydrophobic or basic amino acids in the P1 position, whereas the majority of non-substrates contain Asp or Gln in this position.