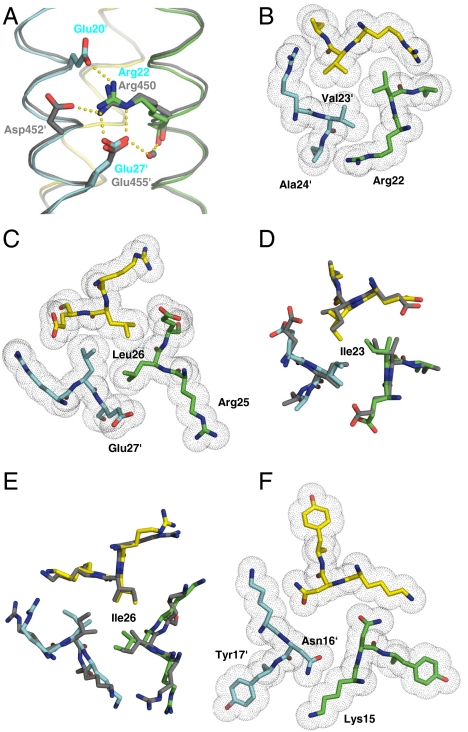
Fig. 4.
Packing of side chains within X-ray crystal structures of GCN4-pM3 and GCN4-pM7. (A) Side view of an overlay of the salt-bridge network (indicated by yellow dots) of the trimerization motifs seen in GCN4-pM3 and the coiled-coil domain of coronin-1 (8) (dark gray ribbons). (B) Axial view of the a4 layer of GCN4-pM3 showing the shielding of the Val23′ residues from solvent by the aliphatic side-chain moieties of Arg22. (C) Axial view of the d4 layer of GCN4-pM3 showing the hydrophobic packing of Ile26 residues and the aliphatic side-chain moieties of the Glu27′. (D and E) Axial views of an overlay of the acute packing of isoleucine residues at the a4 and d4 layers seen in GCN4-pM7 and GCN4-pII (dark gray) (12). (F) Axial view of the a3 layer showing the conformation of the Asn16 residues and the unfavorable packing of their polar moieties against the aliphatic portion of Lys15′ as well as the formation of a large cavity in the hydrophobic core of GCN4-pM3. A similar conformation is seen in the crystal structure of GCN4-pM7. Side chains of residues are shown as stick representation (A–F) and van der Waals spheres (B, C, and F). Monomers are shown as Cα traces and the bound water molecules as a small red (GCN4-pM3) or gray (coiled-coil domain of coronin-1) sphere (A). Oxygen and nitrogen atoms are colored in red and blue, and carbon atoms are shown in green, cyan, and yellow for GCN4-pM3 and GCN4-pM7 monomers A, B, and C, respectively. The prime indicates a residue of chain B.