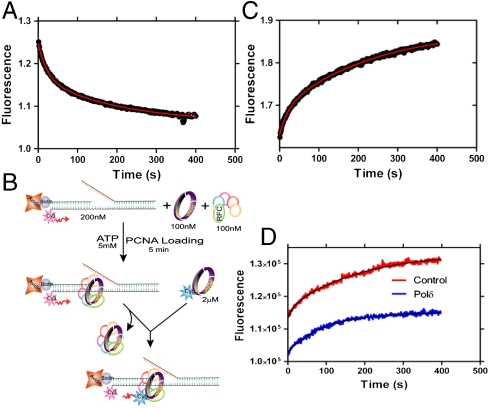
Fig. 3.
Unloading of PCNA. (A) PCNA (100 nM) loaded onto DNA (100 nM) was mixed with unlabeled trap PCNA at 2 μM concentration and the FRET signal was followed at 665 nm. The data were fit to a double-exponential equation (0.048 ± 0.004 s-1 and 0.005 ± 0.001 s-1). (B) Schematic presentation of experimental procedure for Fig. 3C. (C) Experiment performed as depicted in Fig. 3B. RFC loads Cy5-labeled PCNA at a rate equal to its off-rate from DNA. (D) A preassembled complex of unlabeled PCNA (100 nM) with RFC (100 nM) and DNA (200 nM) was mixed with Cy5-labeled PCNA (2 μM) in the absence and presence of polδ (100 nM). In the absence of polδ (control) the traces were fitted to a double-exponential equation with a fast rate constant 0.06 ± 0.005 s-1 and a slow rate constant 0.01 ± 0.003 s-1. In the presence of polδ, the traces were fitted to a single-exponential equation with a rate constant 0.01 ± 0.003 s-1. The amplitude of the signal with polδ is 60.5 ± 0.5% of that observed for the control.