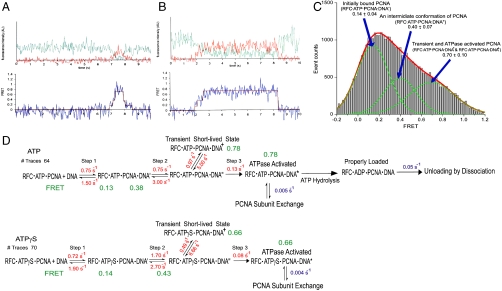
Fig. 4.
Intermediate steps and rates during the process of the PCNA loading. (A and B) Single-molecule fluorescence intensity and FRET time traces showing various types of transitions between the intermediate steps in the PCNA loading process. The traces [fluorescence intensity (Upper) and FRET (Lower)] shown here are from PCNA loading in the presence of ATP. The green line in the intensity trace represents Cy3 fluorescence intensity and the red line represents Cy5 intensity. (C) FRET efficiency histogram of PCNA loading on the substrate DNA in the presence of RFC and ATP or ATPγS. Parts of FRET traces showing nonzero FRET efficiency, selected by visual inspection, were used to construct the histogram. At least three nonzero FRET states are apparent from the histogram as seen in the sample traces. Three Gaussian curves were used to fit the histogram. The lowest FRET state around 0.14 corresponds to the initially bound state of PCNA on the DNA (RFC·ATP·PCNA·DNA′ in Fig. 4D). The middle FRET state around 0.40 corresponds to the next intermediate conformation of PCNA on DNA (RFC·ATP·PCNA·DNA′′ in Fig. 4D). The highest FRET state around 0.70 includes the properly loaded PCNA conformation, improperly loaded PCNA conformation, and the other two transient conformations (RFC·ATP·PCNA·DNA+ and RFC·ATP·PCNA·DNA) as shown in Fig. 4D. (D) Kinetics of PCNA loading acquired from single-molecule (depicted in red) and ensemble (depicted in blue) studies in the presence of ATP or ATPγS. The model consists of FRET states in series where transitions take place only between adjacent states. The states (RFC·ATP·PCNA + DNA and RFC·ATPγS·PCNA + DNA) include an initial bound state RFC·PCNA on DNA (zero FRET), two intermediate states, RFC·PCNA·DNA′ and RFC·PCNA·DNA′′ (medium FRET), and a loaded state (high FRET). The latter consists of the transient species RFC·PCNA·DNA+, ATPase activated state, properly loaded state, and improperly loaded state as implicated by the PCNA unloading ensemble kinetics results. Unloading of PCNA takes place by dissociation or subunit exchange.