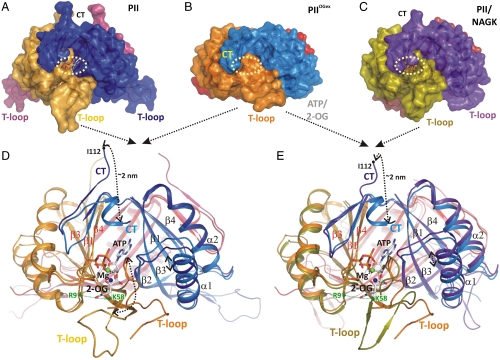
Fig. 1.
Structural basis of PII regulation by 2-oxoglutarate. (A–C) Side views of different S. elongatus PII structures in surface representation. The location of the ATP-binding site is indicated by a yellow dashed circle; the red boxes highlight the structure part discussed detailed in D. (A) Ligand-free PII modeled together with ATP (placed according to the superimposed 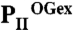 structure). The subunits are color coded in dark blue, light orange, and warm pink. The extended C termini and T loops of the proteins chains are marked CT and T-loop, respectively. (B) PII/ATP/2-OG (
structure). The subunits are color coded in dark blue, light orange, and warm pink. The extended C termini and T loops of the proteins chains are marked CT and T-loop, respectively. (B) PII/ATP/2-OG (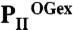 ) complex structure; subunits color coded in orange, marine blue, and red. The structure adopts a compact conformation because of the back folding of the C terminus (CT) onto the core domain structure and the T loop observed in a partially disordered conformation. (C) Structure of PII in the PII–NAGK complex (PDB ID code: 2V5H) modeled together with ATP. Subunits are color coded in magenta, deep olive, and salmon. (D) Superposition of the ligand-free PII and
) complex structure; subunits color coded in orange, marine blue, and red. The structure adopts a compact conformation because of the back folding of the C terminus (CT) onto the core domain structure and the T loop observed in a partially disordered conformation. (C) Structure of PII in the PII–NAGK complex (PDB ID code: 2V5H) modeled together with ATP. Subunits are color coded in magenta, deep olive, and salmon. (D) Superposition of the ligand-free PII and 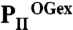 structure as ribbon models in the color codes derived from A and B. Secondary structure elements are marked with β1–β5 and α1–α2 and ATP and 2-OG are shown in stick representation, whereas the Mg ion is marked in green. The sites of two mutations, R9L and K58M, introduced to prove the binding mode of 2-OG are marked with green dots. Remarkable conformational transitions are marked with dashed lines: movement of the C terminus, a shift of helix 1, and displacement of the T-loop base by 2-OG while the tip becomes disordered. (E) Superposition of
structure as ribbon models in the color codes derived from A and B. Secondary structure elements are marked with β1–β5 and α1–α2 and ATP and 2-OG are shown in stick representation, whereas the Mg ion is marked in green. The sites of two mutations, R9L and K58M, introduced to prove the binding mode of 2-OG are marked with green dots. Remarkable conformational transitions are marked with dashed lines: movement of the C terminus, a shift of helix 1, and displacement of the T-loop base by 2-OG while the tip becomes disordered. (E) Superposition of 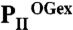 and PII–NAGK structures as ribbon models in the color codes derived from B and C. Structural details are indicated as in D.
and PII–NAGK structures as ribbon models in the color codes derived from B and C. Structural details are indicated as in D.