Abstract
Protein synthesis in all living organisms occurs on ribonucleoprotein particles, called ribosomes. Despite the universality of this process, eukaryotic ribosomes are significantly larger in size than their bacterial counterparts due in part to the presence of 80 r proteins rather than 54 in bacteria. Using cryoelectron microscopy reconstructions of a translating plant (Triticum aestivum) 80S ribosome at 5.5-Å resolution, together with a 6.1-Å map of a translating Saccharomyces cerevisiae 80S ribosome, we have localized and modeled 74/80 (92.5%) of the ribosomal proteins, encompassing 12 archaeal/eukaryote-specific small subunit proteins as well as the complete complement of the ribosomal proteins of the eukaryotic large subunit. Near-complete atomic models of the 80S ribosome provide insights into the structure, function, and evolution of the eukaryotic translational apparatus.
Keywords: homology modeling, RNA, translation, flexible fitting, molecular dynamics
Protein synthesis occurs on large macromolecular complexes, called ribosomes (1). Ribosomes are composed of two subunits, both of which are built from protein and RNA. Bacterial ribosomes, for example, in Escherichia coli, contain a small subunit composed of one 16S rRNA and 21 ribosomal proteins (r proteins), and a large subunit containing 5S and 23S rRNAs and 33 r proteins. In contrast, eukaryotic ribosomes are much larger and more complex, containing additional RNA in the form of so-called expansion segments (ES) as well as many additional r proteins and r-protein extensions. The additional r proteins present in eukaryotic ribosomes are likely to reflect the increased complexity of translation regulation in eukaryotic cells (2–5). Moreover, many of these eukaryote-specific components have been associated with human disorders (4). Thus, structural insight into the localization of these elements will be important to furthering our understanding of eukaryotic translation regulation as well as disease.
Compared with the ∼54 r proteins of the bacterial ribosome, plant and fungal 80S ribosomes contain ∼80 r proteins (see Table S1 for r-protein nomenclature). Crystal structures have revealed the location of each small and large subunit r protein within bacterial ribosomes (6–12) as well as the r proteins within the archaeal large ribosomal subunit (13, 14). In contrast, the localization of ribosomal proteins within eukaryotic 80S ribosomes has come mainly from early studies using immuno-EM and cross-linking approaches (see, for example, refs. 15–18). Moreover, the first molecular models for the eukaryotic ribosome were built at 15-Å resolution by docking the structures of the bacterial small 30S subunit (6) and archaeal large 50S subunit (13), thus only identifying the location of a total of 46 eukaryotic r proteins with bacterial or archaeal homologues (19). Recently, cryo-EM reconstructions of plant and fungal 80S ribosomes have led to the localization of three eukaryote-specific r proteins: RACK1 (20) and S19e (21) in the small subunit and L30e in the large subunit (22). Therefore, the current locations of 49 (33 large and 16 small subunit) r proteins are known for the eukaryotic 80S ribosome, whereas 31 (14 and 17, respectively) remain to be elucidated.
Here we have utilized cryo-EM maps of yeast and wheat germ ribosomes at 5.5 Å (see accompanying article in this issue of PNAS) and 6.1-Å resolution, respectively, to identify the location and build models for 74 of the 80 r proteins in the eukaryotic 80S ribosome, including 12 archaea/eukaryote-specific r proteins in the small subunit and 15 in the large subunit. Near-complete models for the yeast and wheat germ 80S ribosome will be an important resource for researchers working with these model organisms.
Results and Discussion
Placement of Ribosomal Proteins into a 5.5-Å Cryo-EM Map of an 80S Ribosome.
Subtraction of the density assigned to the rRNA (gray in Fig. 1) in the 5.5-Å resolution cryo-EM structure of the Triticum aestivum translating 80S ribosome (see accompanying article in this issue of PNAS) left density that was attributed to r proteins (green in Fig. 1A). Due to the lack of complete sequence information for T. aestivum, sequences of the closely related Oryza sativa were used where necessary (Tables S1–S6). This is a valid approach because of the given resolution of the map and the very high similarity of the proteins (> 90% identity on average). Models for 44 of the 80 r proteins of the T. aestivum 80S ribosome were built into this map using the templates present in the bacterial and archaeal ribosome structures (23, 24). Similarly, 44 of 79 r proteins of the yeast 80S ribosome were built into the previously reported cryo-EM structure of a translating Saccharomyces cerevisiae 80S ribosome at 6.1-Å resolution (25). The archaeal/eukaryote-specific r-protein extensions were modeled de novo whenever possible, building out from N and C terminus of the template-based core regions using electron density map and secondary structure prediction constraints.
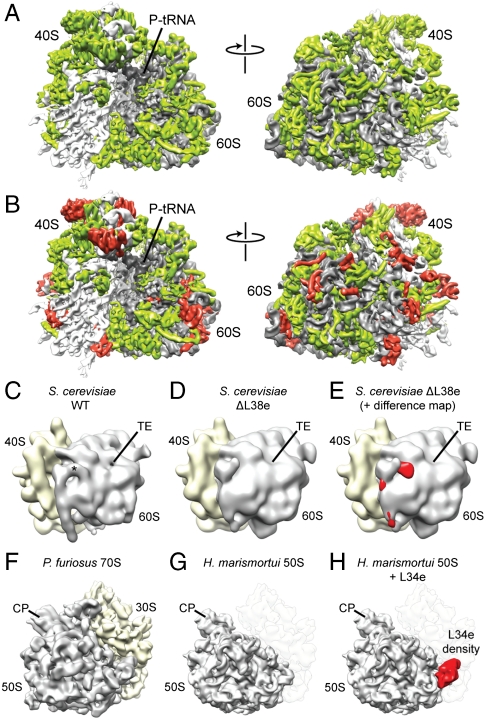
Fig. 1.
Identification of r-proteins L38e and L34e. (A) Cryo-EM map of the T. aestivum 80S ribosome, with rRNA colored gray and r protein colored green. (B) Same as A, but with localized r proteins colored red. Reconstruction of (C) S. cerevisiae WT 80S ribosome compared to (D) reconstruction of S. cerevisiae 80S ribosomes isolated from a strain lacking the gene for L38e. The asterisk indicates the position of additional density assigned to L38e, and the tunnel exit (TE) is shown for reference. (E) Difference density map calculated between C and D and shown superimposed on the map from D. Reconstruction of (F) P. furiosus 70S ribosome, compared to (G) X-ray structure of the 50S subunit from H. marismortui filtered to a similar resolution. (H) Difference density map calculated between F and G and shown superimposed on the map from G identifying the location of r-protein L34e (red).
A total of 17 r proteins (see Table S1 for r-protein family nomenclature), 7 (S4e, S17e, S19e, S24e, S27e, S28e, and RACK1) from the 40S subunit, and 10 (L4e, L6e, L14e, L18ae, L27e, L30e, L35ae, P0, P1, and P2) from the 60S subunit were modeled using available X-ray and NMR structures of free r proteins (Tables S2–S5). Homology models for six r proteins (S25e, L22e, L29e, L34e, L36e, and L38e) were built using HHpred (26) and Modeller (27) on the basis of similarity with domains of proteins of known structure, for example, S25e and L38e were predicted to have helix-turn-helix and K-homology domains, both of which are known to interact with RNA. Seven r proteins (S7e, S21e, S26e, S30e, L13e, L28e, and L41e) were tentatively modeled ab initio on the basis of secondary structure predictions and density characteristics, and six small subunit r proteins (S3ae, S6e, S8e, S10e, S12e, and S27a) could not be localized and were therefore not modeled. The protein models were initially fitted as rigid bodies, merged with the rRNA models and an extended version of molecular dynamics flexible fitting (MDFF) was applied to remove clashes, impose stereochemical restraints, and improve the overall fit to the map (28). At the given resolution, it should be noted that the degree of accuracy and reliability of the assignments varies for the different r proteins (Table S6): The fold and location of ribosomal core proteins and those modeled on the basis of available X-ray and NMR structures will have a higher degree of accuracy than those generated using remote homology or ab initio modeling. Although the latter models can only be considered tentative placements, the location of the r protein is more certain, being consistent with available biochemical evidence (Table S6). Collectively, a total of 74 r proteins were modeled, 27 (excluding P0, P1, P2) of which are not present in the bacterial or archaeal ribosome crystal structures (red in Fig. 1B).
Localization of Ribosomal Proteins of the 80S Ribosome.
The main basis for the localization of r proteins in the cryo-EM reconstructions of the 80S ribosomes was the excellent agreement between the density features in the maps with distinctive protein-fold characteristics of the X-ray structures and homology models (Fig. S1). Additional supporting information was utilized for the localization of r proteins, particular those modeled ab initio. The supporting data included species-specific differences in length between r proteins of wheat germ, yeast, and archaeal ribosomes, as well as the wealth of data available on the spatial arrangement of r proteins in eukaryotic ribosomes derived from a variety of different approaches: (i) the order of assembly of r proteins (29); (ii) accessibility of particular r proteins to proteolysis; (iii) cross-linking of r proteins (15, 18, 30, 31); and (iv) immuno-EM studies (16, 32) (see Table S6). Furthermore, the localization of r-protein L38e was supported by comparison of a cryo-EM reconstruction of wild-type yeast 80S ribosome with that of a yeast 80S ribosome isolated from a strain lacking the gene for r-protein L38e (Fig. 1 C–E). Similarly, comparison of a cryo-EM reconstruction of a 70S ribosome from the archaeon Pyrococcus furiosus with the crystal structure of the large subunit of Haloarcula marismortui led to the localization of r-protein L34e (Fig. 1 F–H). Both L34e and L38e stabilize different conformations of ES27 (see accompanying article in this issue of PNAS). R-protein L28e is not present in the S. cerevisiae genome, and therefore the localization of L28e was possible by generating difference maps between yeast and T. aestivum ribosomes (Fig. S2).
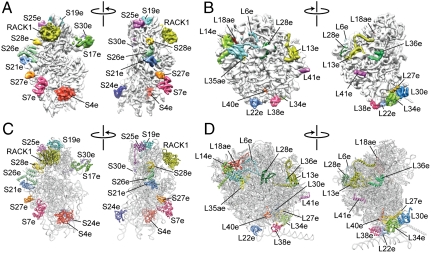
On this basis, it was possible to localize and model a total of 27 r proteins (excluding P0, P1, and P2) that are not present in the crystal structures of bacterial or archaeal ribosomes (Fig. 2 A–D). This encompasses 12 small subunit r proteins (S4e, S7e, S17e, S19e, S21e, S24e, S25e, S26e, S27e, S28e, S30e, and RACK1) and 15 large subunit r proteins (L6e, L13e, L14e, L18ae, L22e, L27e, L28e, L29e, L30e, L34e, L35ae, L36e, L38e, L40e, and L41e) (Fig. 2 A–D). We can assign the unidentified protein interaction partner of RACK1 (33) as being the eukaryote-specific C-terminal extension of r-protein S2p, whereas the localization of L30e on the 60S subunit is as reported for yeast and T. aestivum (Fig. 2 B and D) (22, 34). Mutations in S19e found in Diamond-Blackfan anemia (DBA) patients are clustered around α3 (35), which is seen to interact with h41 in the T. aestivum and yeast 80S models. DBA is an inherited bone marrow failure syndrome that results from defects in ribosomal assembly (4). The localization of S19e (and S28e) to the head of the 40S subunit (Fig. 2A) is also consistent with biochemical data examining assembly precursor particles formed in vivo (29). In addition to S19e, we have localized the other major r proteins associated with DBA, such as S7e on the platform at the base of ES6, S17e to the beak of the 40S subunit, as well as S24e at the interface side bridging h8 and h44 (Fig. 2A).
Fig. 2.
Localization of eukaryote-specific r proteins. Cryo-EM maps of the T. aestivum (A) 40S and (B) 60S subunit, with density for the newly identified r proteins colored distinctly. Molecular models of r proteins of the T. aestivum (C) 40S and (D) 60S subunit, with newly identified r proteins colored distinctly.
Functional Roles for Eukaryote-Specific Ribosomal Proteins.
Although the active sites of the ribosome—the decoding site on the small subunit and the site of peptide-bond formation on the large subunit—are composed largely of rRNA, they are not completely devoid of r proteins (Fig. 3 A–D). Compared with bacterial 30S subunits, eukaryotic 40S subunits contain two additional r proteins, S25e and S30e, with extensions that reach into the decoding and tRNA binding sites (Fig. 3 A and B). Consistent with this localization, S30e has been cross-linked to the 4-thiouridine containing UGA stop codon of mRNA positioned at the A-site (30). Additionally, the C terminus of r-protein S4p is relocated in eukaryotes, due to corresponding rearrangements in h16/17, and reaches from the globular domain on the solvent side right into the decoding site of the small subunit (Fig. 3A). Thus, together with the extensions and loops of eukaryotic homologues to the bacterial S7, S9, S11, S12, and S13 r proteins (11, 12), at least seven different r proteins can interact and modulate the binding of tRNAs to the 40S subunit. At the peptidyl-transferase center on the large subunit, direct interaction is observed between the loop of r-protein L10e and the CCA-end of a peptidyl-tRNA at the P site (Fig. 3C). Based on our model, the loop of L10e is now the r-protein region that comes closest (∼16 Å) to the site of peptide-bond formation (Fig. S3). This loop was disordered and not modeled in the crystal structures of the archaeal 50S subunit (13) and is absent in the bacterial homologue, L16. Instead, the N-terminal extension of the r-protein L27 occupies a similar but distinct position in bacterial ribosomes (36, 37) (Fig. 3D). The loop of L10e is highly conserved and mutations or deletions in this loop are lethal (38), suggesting that it may play an important role in tRNA positioning, as proposed for the N terminus of L27 (36, 37).
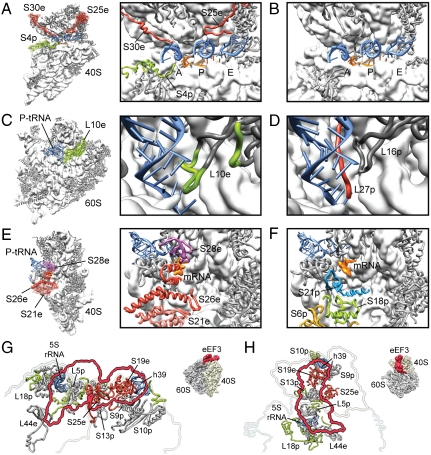
Fig. 3.
Functional roles for eukaryote-specific r proteins. (A) Small 40S subunit with newly modeled r-proteins S30e and S25e (red) and eukaryote-specific extension of S4p (green) highlighted (thumbnail, Left; zoom, Right). (B) Comparative view of the bacterial 30S subunit decoding site (11, 12). In A and B, the anticodon-stemloops of A-, P- and E-tRNAs (blue) and mRNA (orange) are shown for reference. (C) Large 60S subunit with eukaryote-specific extension of L10e (green) highlighted (thumbnail, Left; zoom, Right). (D) Comparative view of the bacterial 50S subunit with bacterial-specific L27p colored red (11). In C and D, the acceptor-stem of the P-tRNA (blue) is shown for reference. (E) Small 40S subunit with newly modeled r-proteins S21e, S26e, and S28e colored distinctly (thumbnail, Left; zoom, Right). (F) Comparative view of the bacterial 30S subunit with bacterial-specific S18p shown in green (11). In E and F, the P-tRNA (blue) and mRNA (orange) are shown for reference. (G and H) The binding site of eEF3 on the S. cerevisiae 80S ribosome, with (G) side and (H) top views (see insets) showing the binding site of eEF3 as a red outline and molecular models of ribosomal components that comprise the eEF3 binding site. Newly identified proteins are shown in red (S19e, S25e) and newly modeled r-protein extensions in green, whereas core r proteins are colored gray. Modified from ref. 48.
Three eukaryote-specific r proteins, S21e, S26e, and S28e, were identified at the mRNA exit site between the platform and head of 40S subunit (Fig. 3E). Both S26e and S28e have been cross-linked from positions (-6 and -7/-10, respectively) in the 5′ untranslated region (UTR) of mRNA (18). The equivalent region of bacterial 30S subunits is occupied by bacterial-specific r proteins S6, S8 as well as S21 in E. coli (6, 10) (Fig. 3F). These differences may reflect the distinct elements found in the 5′ UTRs of bacterial and eukaryotic mRNAs, as well as the divergence in the translation initiation phase (2). For example, eIF3, which is absent in bacteria, interacts with this region of the eukaryotic 40S subunit (32, 39–41). Internal ribosome entry site (IRES) elements present in the 5′ UTR of viral mRNAs also interact with this region of eukaryotic ribosomes (42–45). Indeed, the unknown rpSx that interacts with cricket paralysis virus (CrPV) IRES (45) can now be assigned as r-protein S25e, consistent with a cross-link from the conserved domain 2 fragment from CrPV and other IRESs to S25e (31).
The translation factor binding site is highly conserved on bacterial and eukaryotic ribosomes, with the exception of the aforementioned extensions of r proteins S4p and S30e that reach into the decoding site in the 80S ribosome. Extensions of both S4p and S30e would be expected to interact with domain IV of eEF2, as visualized previously by cryo-EM (46, 47). Additionally, we can now identify the eEF3 interaction partners in the yeast 80S, previously assigned as rpSX1 and rpSX2 (48), as being r-protein S19e and S25e, respectively, both of which are located in the head of the 40S subunit (Fig. 3 G and H). In addition, r-protein L44e as well as eukaryote-specific extensions of r proteins L5p and L18p located within the central protuberance of the 60S subunit also comprise the eEF3 binding site (Fig. 3 G and H).
Coevolution of rRNA Expansion Segments and Eukaryotic-Specific Ribosomal Proteins.
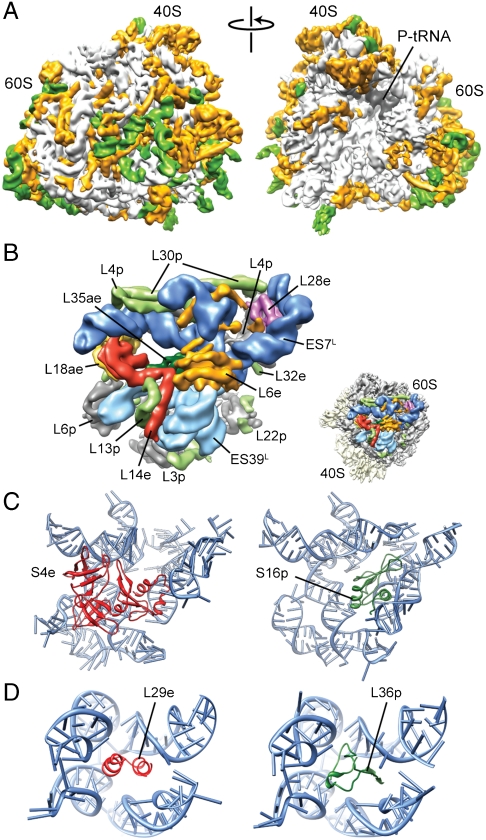
Eukaryotic 80S ribosomes are significantly larger than their bacterial counterparts, the T. aestivum ribosome contains 1.53 MDa (0.62 MDa/40S and 0.91 MDa/60S) of r protein and 1.74 MDa (0.56 MDa/40S and 1.18 MDa/60S) of rRNA, thus totaling 3.27 MDa, whereas E. coli 70S ribosomes total to ∼2.5 MDa (0.9 MDa/30S and 1.6 MDa/50S). Fig. 4A shows that the ES and additional r proteins/protein extensions (green and gold, respectively) form an intricate layer of additional RNA-protein mass that locates predominantly to the solvent surface of the ribosome. The intertwined nature of the additional rRNA ES and r proteins supports the idea that they are coevolving together (49), which is exemplified by the large mass found on the back of the 60S subunit comprising ES7L, ES39L, and five eukaryotic r proteins (L6e, L14e, L18ae, L28e, and L35ae) (Fig. 4B). Interestingly, L6e, L14e, and L27e all adopt the same SH3-like barrel fold, possibly reflecting their origin due to gene duplication events. L27e is located below the L1 stalk on the opposite side of the ribosome from L6e and L14e, where it is sandwiched between H55 and H58. L27e and L34e overlap the position of H58 in the E. coli 70S ribosome, emphasizing the conformational rearrangements that relocate H58 in archaeal/eukaryotic compared to bacterial ribosomes. In contrast, r proteins, such as L13e, L22e, and L36e, occupy empty sites in the bacterial and archaeal ribosomes yet interact with the core rRNA. Interestingly, the loop of H57, which is the interaction partner for L22e, is conserved in eukaryotes, but not in bacteria, which lack this protein.
Fig. 4.
Coevolution of rRNA expansion segments with r proteins in the 80S ribosome. (A) Cryo-EM map of the T. aestivum 80S ribosome, with rRNA ES and variable regions colored green and eukaryote-specific r proteins and extensions colored orange. (B) View of the intertwined region of ES7L (dark blue) and ES39L (light blue), with core r proteins (gray), eukaryote-specific r-protein extensions (pale green), and r proteins (L6e, orange; L14e, red; L18ae, yellow; L28e, pink; L35ae, green) highlighted. Inset shows relative position to 40S (yellow) and 60S subunits (gray). (C) Comparison of relative positions of S4e (red) in yeast/T. aestivum 80S (Left) with S16p (green) in bacteria (11) (Right). (D) Comparison of relative positions of L29e (red) in yeast/T. aestivum 80S (Left) with L36p (green) in bacteria (11) (Right).
Evolution of the Eukaryotic Ribosome.
A previous comparison of archaeal and bacterial large subunits illustrated examples of potential convergent evolution, where evolutionarily unrelated r proteins have evolved to stabilize the same region of 23S rRNA (14). Many such examples are also found by comparing the models of the yeast and T. aestivum 80S ribosome with the archaeal and bacterial crystal structures: The N-terminal domain of S4e overlaps the binding position of S16p (Fig. 4C), and the extended N terminus of L32e overlaps regions of bacterial-specific r proteins L20p and L21p. Likewise, L18ae has two ubiquitin-like α/β roll domains (ULDs), with the N-terminal ULD overlapping bacterial L25p, and like L25p also interacting with the 5S rRNA, whereas α-helix 1 of the C-terminal ULD inserts in the minor groove of H41. Furthermore, L29e sits in a small RNA pocket at the stalk base, which is occupied by L36p in bacteria (Fig. 4D). The localization of L29e to this pocket was based partly on the observation that the stalk rearranges position to establish contact with the head of the 40S subunit in a reconstruction of the yeast ΔL29e-80S (Fig. S4), which has not been observed in any previous yeast 80S reconstructions. Moreover, the assigned position for L29e is in close proximity to L10e (L16p), which exhibits synthetic lethality with L29e in yeast (50).
Conclusion
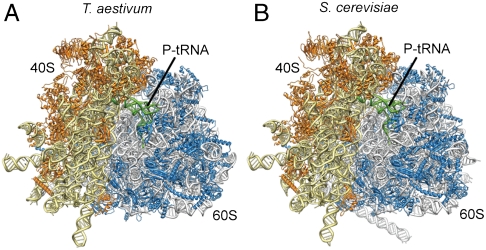
Molecular models are presented for translating T. aestivum and yeast 80S ribosomes encompassing ∼98% of the rRNA and 92.5% of the r proteins (Fig. 5). Given that mammalian ribosomes have the same complement of 80 r proteins as those of T. aestivum presented here, we believe that the information gained from the T. aestivum and yeast 80S models should thus not only provide a resource for researchers working with these model organisms, but may also provide useful information when studying mammalian systems.
Fig. 5.
Structures of wheat germ and yeast eukaryotic 80S ribosomes. (A and B) Near-complete molecular models for the (A) T. aestivum and (B) S. cerevisiae 80S ribosome, with rRNA and protein shown in yellow and orange for the small subunit and gray and blue for the large subunit, respectively.
Experimental Procedures
Sample Preparation and Cryoelectron Microscopy.
Yeast (Saccharomyces cerevisiae) 80S ribosomes were isolated from wild-type strains and strains lacking the genes encoding r proteins L29e and L38e (29), as described previously (25, 51). Cryo-EM reconstructions of the yeast ΔL29e and ΔL38e-80S ribosomes were performed on a Tecnai G2 Spirit transmission electron microscope at 120 kV at a nominal magnification of 90,000 using an Eagle 4,096 × 4,096 pixel CCD camera (FEI) resulting in a pixel size of 3.62 Å/pixel. For the final yeast ΔL29e and ΔL38e-80S ribosome reconstructions, 7,272 and 10,356 particles were used. The cryo-EM map used for modeling of the yeast 80S ribosome was published previously (Electron Microscopy Data Bank ID 1669; ref. 25). Cryo-EM reconstructions of the P. furiosus 70S ribosomes were performed as for the T. aestivum ribosome nascent chain complex samples described in the accompanying article in this issue of PNAS. The final reconstruction of the P. furiosus 70S ribosome used 54,979 particles, yielding a final contrast transfer function corrected map at a resolution of 10 Å. Densities for the 40S subunit, the 60S subunit, and the P-site tRNA were isolated using binary masks.
Density Map Sharpening.
For modeling of r-protein extensions, density maps were sharpened using a nonnegative deconvolution method (Hirsch, Schölkopf and Habeck, accepted) based on the multiplicative updates proposed in (52). As a blurring function, an isotropic Gaussian kernel (generated with the EMAN software package command pdb2mrc for a Protein Data Bank file containing a single atom) was chosen. In addition, a nonnegative background density was introduced to account for solvent contributions and other artefacts. The background was constrained to be uncorrelated with the deconvolved density map. Both the deconvolved map and the background density were then estimated simultaneously using interleaved multiplicative updates. The deconvolution algorithm was run for different kernel sizes and constraint strengths. The most informative density map was selected by visual inspection (Fig. S1).
Homology Modeling of R Proteins.
Based on the crystal structures of the archaeal 50S subunit (13) and the bacterial ribosomal structures (10, 11), it was possible to generate S. cerevisiae and T. aestivum (or O. sativa) homology models (Tables S2–S5). In addition, there are also 12 structures of r proteins obtained from either X-ray or NMR structures in a non-ribosome-associated state (Tables S2–S5). The best templates were chosen by screening available structures and selecting on the basis of both the sequence identity and fitting to the cryo-EM density. Sequence to structure matching has been performed based on profile–profile alignments (53, 54). Alignments were performed using a number of alignment servers, including ClustalW (55), TCoffee (56), MUSCLE (57), and Mafft (58). Using Modeler (59), numerous models were created and ranked based on the discrete optimized protein energy (60) score. From the top scoring models, two were chosen and rigidly fitted into the EM density using Chimera (61) and Coot (62), and the best fit was taken for further refinement. Extended parts of the proteins that did not have a template were truncated at this step and manual adjustments were introduced to the rigidly fitted protein to best fit the density. Whenever modeling of extensions appeared possible on the basis of information in the cryo-EM map, secondary structure predictions were performed (63), along with search for an appropriate template among existing structures using HHpred (64). This information, together with the density information in close proximity to the protein core was examined and, if possible, the extended part was modeled. In cases of ambiguous density, comparison of additional maps (S. cerevisiae, deconvolved T. aestivum; see Fig. S1) was used. Using this approach, a total of over 2,000 amino acids were modeled de novo. The increasing number of modeled extensions allowed us to iteratively minimize the amount of available density, thus providing constraints to find additional solutions to RNA and protein localization.
Refinement and Fitting of the R Proteins into the EM Densities.
Because common methods for protein modeling are to date not capable of incorporating EM data or interaction with RNA directly in the modeling process, the proteins still needed to be flexibly fitted into the density and reconciled with RNA models. Thus, subsequent to the fitting and modeling of the rRNA, proteins were introduced in the model using visual molecular dynamics (VMD) (65), and interactive MDFF was used to refine the proteins into the density using default parameters (28). In regions where the protein density was weak, the location of protein regions was determined by visual inspection, and harmonic constraints to the alpha carbons of those regions were imposed to preserve such location. This process resulted in a rearrangement of the proteins to fit the density, and to resolve protein–RNA and protein–protein clashes while preserving secondary structure. Further MDFF refinement was then performed on the entire 80S model. The fitting was performed iteratively, starting with the most reliable fits, such as docking of X-ray structures and homology models of r proteins. This reduced the leftover density available for localization and modeling of the remaining unassigned r proteins or r-proteins extensions, that later underwent further refinement.
Visualization and Figure Preparation.
Cryo-EM maps and models were visualized and all figures were generated using VMD (65), Chimera (61), and/or PyMol (http://www.pymol.org).
Supplementary Material
Acknowledgments.
This research was supported by grants from the Deutsche Forschungsgemeinschaft SFB594 and SFB646 (to R.B.), SFB740 (to T.M.), and WI3285/1-1 (to D.N.W.), by the European Union and Senatsverwaltung für Wissenschaft, Forschung und Kultur Berlin (UltraStructureNetwork, Anwenderzentrum), and a Marie Curie International Incoming Fellowship within the seventh European Community Framework Programme (E.V.). Computer time for MDFF was provided by the Leibniz-Rechenzentrum.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Coordinates of the atomic models of yeast and Triticum aestivum 80S complex have been deposited in the Protein Data Bank (PDB), www.pdb.org [PBD ID codes 3IZE, 3IZF, 3IZD (yeast rRNA), 3IZB, 3IZC (yeast r proteins), 3IZ7, 3IZ9 (T. aestivum rRNA), and 3IZ6, 3IZ5 (T. aestivum r proteins)]. The cryoelectron microscopic map of the T. aestivum 80S-RNCs has been deposited in the 3D-Electron Microscopy Data Bank (EMDB, http://www.ebi.ac.uk/pdbe/emdb/) (EMBD ID code EMD-1780).
See companion article on page 19748.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010005107/-/DCSupplemental.
References
- 1.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: Diseases of ribosome biogenesis. Mol Biosyst. 2010;6:481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010;33:173–182. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 7.Schluenzen F, et al. Structure of functionally activated small ribosomal subunit at 3.3 A resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 8.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 9.Harms J, et al. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- 10.Schuwirth B, et al. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 11.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 12.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 13.Ban N, et al. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 14.Klein D, Moore P, Steitz T. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 15.Gross B, Westermann P, Bielka H. Spatial arrangement of proteins within the small subunit of rat liver ribosomes studied by cross-linking. EMBO J. 1983;2:255–260. doi: 10.1002/j.1460-2075.1983.tb01414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marion MJ, Marion C. Localization of ribosomal proteins on the surface of mammalian 60S ribosomal subunits by means of immobilized enzymes. Correlation with chemical cross-linking data. Biochem Biophys Res Commun. 1987;149:1077–1083. doi: 10.1016/0006-291x(87)90518-3. [DOI] [PubMed] [Google Scholar]
- 17.Lutsch G, et al. Immunoelectron microscopic studies on the location of ribosomal proteins on the surface of the 40S ribosomal subunit from rat liver. Eur J Cell Biol. 1990;51:140–150. [PubMed] [Google Scholar]
- 18.Pisarev AV, et al. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spahn CM, et al. Structure of the 80S ribosome from Saccharomyces cerevisiae-tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 20.Sengupta J, et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol. 2004;11:957–962. doi: 10.1038/nsmb822. [DOI] [PubMed] [Google Scholar]
- 21.Taylor DJ, et al. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halic M, et al. Localization and dynamic behavior of ribosomal protein L30e. Nat Struct Mol Biol. 2005;12:467–468. doi: 10.1038/nsmb933. [DOI] [PubMed] [Google Scholar]
- 23.Ban N, et al. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 24.Clemons WMJ, et al. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 25.Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hildebrand A, Remmert M, Biegert A, Soding J. Fast and accurate automatic structure prediction with HHpred. Proteins. 2009;77(Suppl 9):128–132. doi: 10.1002/prot.22499. [DOI] [PubMed] [Google Scholar]
- 27.Eswar N, et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006:5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trabuco LG, et al. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferreira-Cerca S, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 30.Bulygin K, et al. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30, and S15. Biochemistry. 2005;44:2153–2162. doi: 10.1021/bi0487802. [DOI] [PubMed] [Google Scholar]
- 31.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bommer UA, Lutsch G, Stahl J, Bielka H. Eukaryotic initiation factors eIF-2 and eIF-3: Interactions, structure and localization in ribosomal initiation complexes. Biochimie. 1991;73:1007–1019. doi: 10.1016/0300-9084(91)90142-n. [DOI] [PubMed] [Google Scholar]
- 33.Chandramouli P, et al. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao JA, Williamson JR. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure. 2004;12:1165–1176. doi: 10.1016/j.str.2004.04.023. [DOI] [PubMed] [Google Scholar]
- 35.Gregory LA, et al. Molecular basis of Diamond-Blackfan anemia: Structure and function analysis of RPS19. Nucleic Acids Res. 2007;35:5913–5921. doi: 10.1093/nar/gkm626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maguire BA, et al. A protein component at the heart of an RNA machine: The importance of protein l27 for the function of the bacterial ribosome. Mol Cell. 2005;20:427–435. doi: 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Voorhees RM, et al. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofer A, Bussiere C, Johnson AW. Mutational analysis of the ribosomal protein Rpl10 from yeast. J Biol Chem. 2007;282:32630–32639. doi: 10.1074/jbc.M705057200. [DOI] [PubMed] [Google Scholar]
- 39.Westermann P, Nygard O. The spatial arrangement of the complex between eukaryotic initiation factor eIF-3 and 40S ribosomal subunit. Cross-linking between factor and ribosomal proteins. Biochim Biophys Acta. 1983;741:103–108. doi: 10.1016/0167-4781(83)90015-5. [DOI] [PubMed] [Google Scholar]
- 40.Westermann P, Nygaard O. Crosslinking of mRNA to initiation factor eIF-3, 24 kDa cap binding protein and ribosomal proteins S1, S3/3a, S6 and S11 within the 48S pre-initiation complex. Nucleic Acids Res. 1984;12:8887–8897. doi: 10.1093/nar/12.23.8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava S, Verschoor A, Frank J. Eukaryotic initiation factor-3 does not prevent association through physical blockage of the ribosomal subunit-subunit interface. J Mol Biol. 1992;226:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- 42.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 43.Spahn CM, et al. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes; the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Boehringer D, et al. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: Remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Schuler M, et al. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 46.Gomez-Lorenzo MG, et al. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 angstrom resolution. EMBO J. 2000;19:2710–2718. doi: 10.1093/emboj/19.11.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spahn CM, et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 2004;23:1008–1019. doi: 10.1038/sj.emboj.7600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andersen BF, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;433:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 49.Yokoyama T, Suzuki T. Ribosomal RNAs are tolerant toward genetic insertions: Evolutionary origin of the expansion segments. Nucleic Acids Res. 2008;36:3539–3551. doi: 10.1093/nar/gkn224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeLabre ML, Kessl J, Karamanou S, Trumpower BL. RPL29 codes for a non-essential protein of the 60S ribosomal subunit in Saccharomyces cerevisiae and exhibits synthetic lethality with mutations in genes for proteins required for subunit coupling. Biochim Biophys Acta. 2002;1574:255–261. doi: 10.1016/s0167-4781(01)00372-4. [DOI] [PubMed] [Google Scholar]
- 51.Beckmann R, et al. Alignment of conduits for the nascent polypeptide chain in the ribosome- Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- 52.Sha F, Lin Y, Saul LK, Lee DD. Multiplicative updates for nonnegative quadratic programming. Neural Comput. 2007;19:2004–2031. doi: 10.1162/neco.2007.19.8.2004. [DOI] [PubMed] [Google Scholar]
- 53.Eswar N, Madhusudhan MS, Marti-Renom MA, Sali A. PROFILE_SCAN: A module for fold assignment using profile-profile scanning in MODELLER. 2005. http://www.salilab.org/modeller.
- 54.Marti-Renom MA, Madhusudhan MS, Sali A. Alignment of protein sequences by their profiles. Protein Sci. 2004;13:1071–1087. doi: 10.1110/ps.03379804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 57.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 60.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 62.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 63.Biegert A, et al. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Res. 2006;34:W335–339. doi: 10.1093/nar/gkl217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.