Abstract
Although the ability of disulfiram to inactivate CYP2E1 has been known for more than 20 years, the mechanism has not yet been elucidated. A metabolite of disulfiram, diethyldithocarbamate (DDC), is converted by CYP2E1 to a reactive intermediate that subsequently inactivates the protein, leading to mechanism-based inactivation. Mass spectral analysis of the inactivated human 2E1 protein demonstrates that the inactivation is due to the formation of an adduct of the reactive metabolite of DDC with the apoprotein. These data, along with mass spectral analysis of a reactive intermediate trapped with GSH, indicate the involvement of a reactive intermediate with a molecular mass of 116 Da. Our results suggest that this binding involves formation of a disulfide bond with one of the eight cysteines in CYP2E1. The inactivation of wild-type CYP2E1 as well as two of its polymorphic mutants, CYP2E1*2 and CYP2E1*4, was also investigated. For wild-type CYP2E1, the KI was 12.2 μM and the kinact was 0.02 min−1. The KI values for the two polymorphic mutants were 227.6 and 12.4 μM for CYP2E1.2 and CYP2E1.4, and the kinact values were 0.0061 and 0.0187 min−1, respectively. These data indicate that DDC is a much less efficient inactivator of CYP2E1.2 than it is of either the wild-type or the CYP2E1.4 variant.
Introduction
Disulfiram (Antabuse) has been used therapeutically for more than 60 years for the treatment of alcoholism because of its ability to inhibit aldehyde dehydrogenase (ALDH) (Hald and Jacobsen, 1948; Gaval-Cruz and Weinshenker, 2009). This inhibition leads to a buildup of acetaldehyde in those patients who continue to consume alcohol. The elevated levels of acetaldehyde lead to nausea, vomiting, and headaches (Kitson, 1977).
Disulfiram has been shown to have a variety of other therapeutic uses because of its ability to inhibit several other pharmacologically important target enzymes. One of the enzymes inhibited by disulfiram is dopamine β-hydroxylase (DBH), which metabolizes dopamine to norepinephrine (Musacchio et al., 1966). This ability to inhibit the metabolism of dopamine to norepinephrine has resulted in some physicians prescribing disulfiram as a means to promote cocaine abstinence (Gaval-Cruz and Weinshenker, 2009). A third enzyme that is inhibited by disulfiram is glutathione transferase, which is involved in the conjugation of endogenously formed electrophilic compounds with GSH (Ploemen et al., 1996).
Disulfiram inhibits these enzymes by two different mechanisms, both of which are dependent on the bioactivation of disulfiram by the body. The first step in these mechanisms involves the reduction of disulfiram in the blood to its monomer, diethyldithocarbamate (DDC), by the glutathione reductase system in erythrocytes (Cobby et al., 1977). DDC has been shown to be the metabolite of disulfiram responsible for the inhibition of DBH (Goldstein, 1966). Because the catalytic activity of DBH is copper-dependent, the depletion of copper by DDC causes the loss of DBH activity (Goldstein, 1966). The inactivation of ALDH by disulfiram also is a result of the metabolic conversion of DDC by a series of enzymes to ultimately form methyl N,N-diethylthiocarbamoyl sulfoxide. This compound can then undergo N-dealkylation, resulting in the formation of a covalent adduct with ALDH. Formation of this adduct on the cysteine 302 of ALDH is responsible for the inactivation of the catalytic activity (Lipsky et al., 2001).
Another enzyme that is inhibited by the administration of disulfiram is human CYP2E1. CYP2E1 is involved in the metabolism of numerous low-molecular-weight xenobiotics including acetaminophen, benzene, and chlorzoxazone, as well as anesthetics such as halothane (Koop, 1992; Lieber, 1997). Disulfiram has been used for a long time as an inhibitor of CYP2E1 in various in vivo studies (Doroshyenko et al., 2009). It has also been proposed that inhibition by disulfiram could be used to prevent the activation of nitrosamine carcinogens by CYP2E1 (Wattenberg et al., 1989). The inhibition of CYP2E1 by disulfiram has been reported to be due to mechanism-based inactivation of the P450 by an oxidized metabolite of disulfiram formed by the 2E1, which reacts with the enzyme, leading to inactivation (Guengerich et al., 1991). One of the primary reasons that disulfiram is widely used for inhibition of CYP2E1 in human studies is its selectivity. In addition, it is relatively nontoxic, except when the patient consumes alcohol. Studies have shown that disulfiram has no effect on human CYP2C9, 2C19, 2D6, or 3A4 (Kharasch et al., 1999).
Although the ability of disulfiram to act as a mechanism-based inactivator of CYP2E1 has been known for 20 years, the exact mechanism by which it causes this inactivation has not been elucidated. We report here that disulfiram by itself does not inactivate CYP2E1 in an in vitro reaction; however, its reduced form, DDC, does inactivate CYP2E1 through the formation of a covalent adduct with CYP2E1. We have identified the metabolite of DDC that is responsible for the inactivation of CYP2E1. We have also characterized the kinetics for the inactivation of the wild-type human CYP2E1 as well as two of its commonly occurring polymorphic forms, CYP2E1.2 and CYP2E1.4. These polymorphisms result in the substitution of the arginine residue at position 76 of 2E1 by a histidine and the valine at position 179 by an isoleucine. These two amino acid substitutions have been found in white and Chinese populations with frequencies estimated to be between 2.4 and 2.6% (Hu et al., 1997; Fairbrother et al., 1998).
Materials and Methods
Chemicals.
NADPH, GSH, catalase, dilauroylphosphatidylcholine, disulfiram, and DDC were purchased from Sigma-Aldrich (St. Louis, MO). 7-Ethoxy-4-(trifluromethyl)-coumarin (7-EFC) was obtained from Invitrogen (Carlsbad, CA). All other chemicals and solvents used were of the highest purity available from commercial sources.
Purification of Enzymes.
The plasmid for human CYP2E1 was a generous gift from Dr. James R. Halpert (University of California at San Diego, La Jolla, CA). CYP2E1 and the two variants (*2 and *4) were expressed in truncated forms in which the hydrophobic membrane-spanning domain had been removed (Δ3–21), and, in addition, a His-tag was added to the C terminus. The CYP2E1 proteins were overexpressed in Escherichia coli C41 (DE3) cells and purified to homogeneity as described previously (Scott et al., 2001). Truncated bacterially expressed rabbit CYP2E1 was purified as described previously (Larson et al., 1991; Kent et al., 1998). NADPH-cytochrome P450 reductase and cytochrome b5 were expressed and purified as described previously (Lin et al., 2005).
Site-Directed Mutagenesis of CYP2E1.
The mutations of CYP2E1 were accomplished with the QuikChange site-directed mutagenesis kit according to the manufacturer's protocol (Stratagene, La Jolla, CA). The primers used for the mutagenesis were 5′-CGTGGGCTCGCAGCACATGGTGGTGATGCACGG-3′ and 5′-CCGTGCATCACCACCATGTGCTGCGAGCCCACG-3′ for CYP2E1*2 and 5′-CCAAGGGCACAGTCGTAATACCAACTCTGGACTCTG-3′ and 5′-CAGAGTCCAGAGTTGGTATTACGACTGTGCCCTTGG-3′ for CYP2E1*4. The site-specific mutations were confirmed by DNA sequencing at the University of Michigan Sequencing Core facility.
Enzyme Assay and Inactivation of P450s.
To assess catalytic activity, CYP2E1 (1.2 nmol) was reconstituted with reductase and cytochrome b5 (1:2:2 ratio) with 200 μg/ml dilauroylphosphatidylcholine in a final volume of 800 μl for 1 h on ice. The primary reaction mixtures for the inactivation studies contained 100 pmol of CYP2E1 in the reconstituted system and 10 units of catalase in a final volume of 100 μl of 100 mM potassium phosphate buffer, pH 7.3. Different concentrations of disulfiram or DDC were added to separate primary reactions. These reactions were then initiated by the addition of NADPH to a final concentration of 1.3 mM. At the times indicated, aliquots (15 μl) of the primary reaction mixture were removed and transferred to a secondary reaction mixture containing 0.3 mM NADPH and 0.1 mM 7-EFC in 100 mM potassium phosphate buffer, pH 7.3. The secondary reaction mixtures were incubated for 15 min at 28°C and then the reactions were terminated by the addition of 100 μl of acetonitrile. The activity remaining was determined by measuring the fluorescent intensity of the product at 520 nm with excitation at 410 nm. Kinetic analyses were performed using GraphPad Prism (version 5 for Mac; GraphPad Software Inc., San Diego, CA).
Analysis of the Primary Metabolite of DDC Formed by 2E1 Using HPLC.
CYP2E1 (1.2 nmol) was reconstituted as described above with reductase (2.4 nmol) in a total volume of 800 μl. Reactions were performed in 100-μl aliquots with 100 μM DDC. In control samples, 2.6 μl of water was added, and in the metabolism samples, 2.6 μl of 50 mM NADPH was added to initiate the reaction. All samples were incubated at 28°C for 1 h. At the end of 1 h samples were injected (10 μl) onto a Shimadzu high-performance liquid chromatograph. The HPLC column was a Zorbax C18 reverse-phase column (4.6 × 25 cm, 300 Å; Agilent Technologies, Santa Clara, CA). The solvent system consisted of solvent A (10 mM ammonium formate in water) and solvent B (100% methanol). The column was eluted with 30% B for 15 min, and then a linear gradient from 30 to 95% B was used over 30 min at a flow rate of 0.8 ml/min. The elution was analyzed by a model 996 diode array detector (Millipore Corporation, Billerica, MA).
Analysis of the Modified CYP2E1 Protein by ESI-LC-MS after Inactivation by Sodium Diethyldithiocarbamate.
Rabbit CYP2E1 was reconstituted with reductase in a 1:1 ratio as described above. The rabbit CYP2E1 was then incubated for 30 min with 100 μM sodium diethyldithiocarbamate and 1.3 mM NADPH in 100 mM potassium phosphate, pH 7.3. After incubation, 50 μl of the reaction mixture was injected onto a reverse-phase C3 column (2 × 150 mm, 5 mm; Agilent Technologies). CYP2E1 was separated from other reaction components with a binary solvent system consisting of 0.1% trifluoroacetic acid in water (solvent A) and 0.1% trifluoroacetic acid in acetonitrile (solvent B) using the gradient: 30% B for 5 min, linearly increased to 90% B in 20 min, and held at 90% B for 30 min. The flow rate was 0.3 ml/min. The molecular masses of the unmodified and DDC-modified CYP2E1 were determined by deconvolution of the apoprotein charge envelopes using BioWorks software (Thermo Fisher Scientific, Waltham, MA) (Zhang et al., 2009).
LC-MS/MS Analysis of GSH Conjugate.
Human CYP2E1 was reconstituted as described above and incubated with 10 mM GSH, 100 μM DDC, and 1.3 mM NADPH at 28°C for 30 min. The samples were then added to a 1-ml AccuBOND ODS-C18 solid-phase extraction cartridge (Agilent Technologies) previously washed with 2 ml of methanol followed by 2 ml of water. After the samples were added to the columns, the cartridges were washed with 2 ml of water and then eluted with 2 ml of methanol followed by 0.3 ml of acetonitrile. The samples eluted in the acetonitrile and methanol were dried under N2 gas and resuspended in 70 μl of 50% acetonitrile containing 0.5% acetic acid. The samples were analyzed by LC-MS/MS as described previously (Lin et al., 2009).
Results
Inactivation of Human CYP2E1 in the Reconstituted System by Disulfiram and Diethyldithiocarbamate.
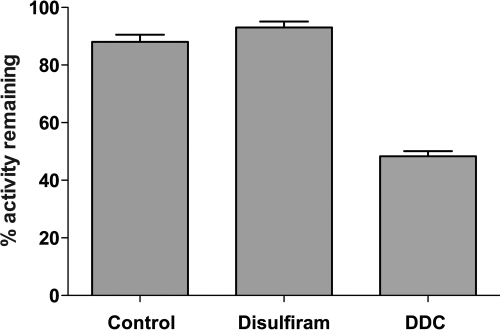
Both disulfiram and DDC have been known for a long time to be inhibitors of CYP2E1 in vivo. The inactivation kinetics of human CYP2E1 by both disulfiram and DDC were investigated in our in vitro reconstituted system. Human CYP2E1 in the reconstituted system was incubated either with control solvent (dimethyl sulfoxide), 20 mM disulfiram, or 200 μM DDC in water. After incubation for 30 min, aliquots were transferred to a secondary reaction mixture containing NADPH and 7-EFC. After 15 min, the reactions were terminated, and the fluorescence was measured for each of the reactions as described under Materials and Methods. As shown in Fig. 1, the CYP2E1 sample containing disulfiram exhibited no loss in activity compared with the control; however, the sample containing DDC demonstrated a significant loss of activity (49%) as has been reported previously (Guengerich et al., 1991). These results demonstrate that disulfiram does not inactivate CYP2E1 in the reconstituted system but that DDC is a good inactivator.
Fig. 1.
Effects of disulfiram and diethyldithocarbamate on human CYP2E1 in the reconstituted system. CYP2E1 was reconstituted with reductase in primary reaction mixtures containing either 20 mM disulfiram or 200 μM DDC. Samples were incubated for 0 or 30 min after the addition of NADPH and then transferred to secondary reaction mixtures containing 0.1 mM 7-EFC and 0.3 mM NADPH. The secondary reactions were incubated for 15 min and terminated by the addition of 100 μl of acetonitrile, and the 7-hydroxy-4-trifluoromethyl coumarin fluorescence was measured as described under Materials and Methods. Percentages are calculated on the basis of the amount of signal for the 30-min incubations divided by the signal for the 0-min incubations from experiments done in triplicate on 2 separate days.
Kinetics of the Mechanism-Based Inactivation of CYP2E1 by Diethyldithocarbamate.
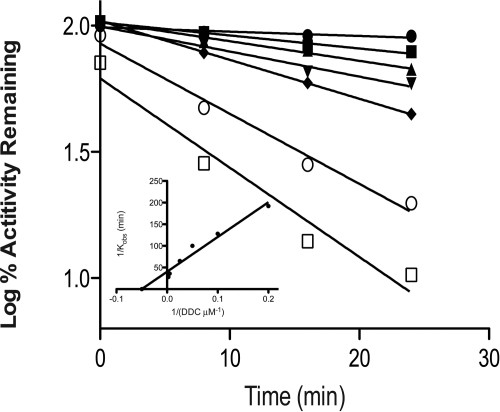
Figure 2 shows the time courses for the inactivation of CYP2E1 at various concentrations of DDC. The inactivation of the 7-EFC O-deethylation activity of CYP2E1 exhibited pseudo-first-order kinetics with respect to time and required NADPH. Linear regression analyses of the time course data were used to determine the initial rate constants for inactivation (kobs) at the various concentrations of DDC. From the double reciprocal plot (Fig. 2) of the values of kobs and the concentration of DDC, the KI was determined to be 12.2 μM and the kinact was 0.02 min−1.
Fig. 2.
Time- and concentration-dependent inactivation of CYP2E1 by DDC. Reconstituted CYP2E1 was incubated with 0 (●), 5(■), 10 (▴), 20 (▾) 40 (♦), 200 (○), and 400 (□) μM DDC. Aliquots were removed at the times indicated and assayed for residual activity as described under Materials and Methods. The inset shows the double reciprocal plot for the initial rates of inactivation as a function of the concentrations of DDC. The kinetic constants were determined from the double reciprocal plot. The data shown represent the average of four separate experiments done in duplicate.
Inactivation of Polymorphic CYP2E1 by DDC.
Several genetic polymorphisms of human CYP2E1 have been reported (Hu et al., 1997). We investigated two that have amino acid substitutions in exons 2 and 4, CYP2E1*2 and CYP2E1*4, respectively. These polymorphisms have been estimated to be present in 2.4 to 2.6% of the white and Chinese populations (Hu et al., 1997; Fairbrother et al., 1998). Previous studies on the catalytic properties of these proteins have demonstrated that CYP2E1.4 possesses properties similar to those of the wild-type CYP2E1.1; however, CYP2E1.2 exhibits kinetic properties significantly different from those of the wild type and they were substrate-dependent (Hanioka et al., 2003).
The inactivations of the two polymorphic CYP2E1 forms were investigated as described above. As shown in Table 1, the KI values for CYP2E1.2 and CYP2E1.4 are 227.6 and 12.4 μM, respectively, and the kinact values are 0.006 and 0.019 min−1, respectively. The values for kinact/KI for these two proteins are 2.6 × 10−6 and 1.5 × 10−3, respectively. Thus, the kinact/KI for CYP2E1.2 was approximately 1000-fold less than that for the wild type, whereas CYP2E1.4 exhibited a relatively small change.
TABLE 1.
Inactivation kinetics for CYP2E1 alleles
P450 (15 pmol) was incubated with DDC in the reconstituted system and the kinetic values were calculated as described under Materials and Methods.
| 2E1 allele | KI | kinact | kinact/KI |
|---|---|---|---|
| μM | min−1/μM | ||
| Human *1 | 12.2 | 0.0202 | 1.6 × 10−3 |
| Human *2 | 228 | 0.0061 | 2.7 × 10−6 |
| Human *4 | 12.5 | 0.0187 | 1.5 × 10−3 |
| Rabbit | 5.8 | 0.0075 | 1.3 × 10−3 |
HPLC Analysis of the Metabolism of DDC.
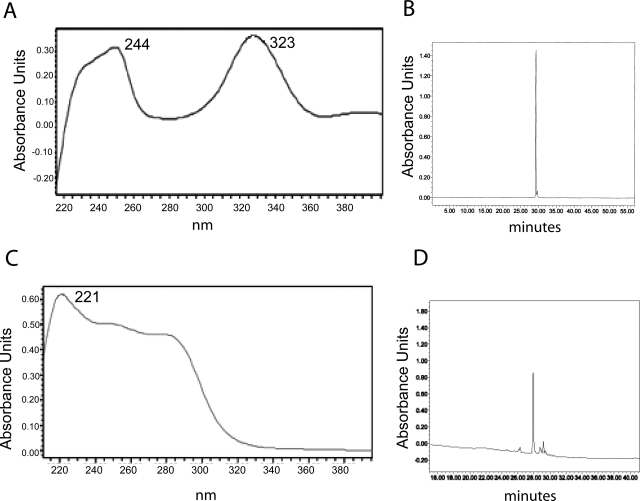
Because the metabolism of DDC by purified CYP2E1 in the reconstituted system has not been reported previously, we investigated DDC metabolism using HPLC. DDC was incubated with reconstituted CYP2E1 in the presence and absence of NADPH. Before metabolism, DDC exhibited a UV spectrum with peaks at 244 and 323 (Fig. 3A), as reported previously (Ploemen et al., 1996), and it eluted as a single peak with a retention time of 29.5 min (Fig. 3B). Incubation of DDC with 2E1 in the reconstituted system with NADPH produced one primary product having a peak at 221.1 in the UV spectrum (Fig. 3C), which eluted at a slightly earlier time of 27.8 min (Fig. 3D).
Fig. 3.
Metabolism of diethyldithocarbamate by CYP2E1. DDC (100 μM) was incubated with reconstituted CYP2E1 (1 pmol/μl) in the reconstituted system. The control and NADPH+ samples were incubated for 30 min at 28°C. Samples were then injected onto the HPLC column, eluted, and monitored as described under Materials and Methods. The control samples (A) exhibited two peaks in the UV spectrum absorbing at 244 and 323 nm and eluting as a single peak having an elution time at 29.5 min (B). The sample incubated with NADPH (C) gave a single major metabolite with a UV spectrum peak at 221.1 (C) and having an elution time of 27.8 min (D).
Covalent Binding of DDC to the CYP2E1 Apoprotein.
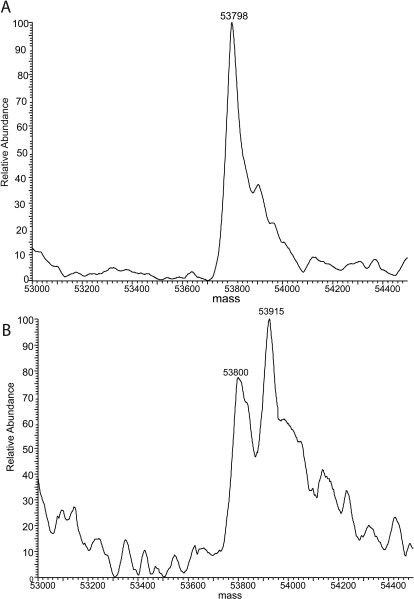
To determine whether the mechanism-based inactivation of CYP2E1 by DDC is due to the formation of adducts of DDC with the protein or the heme, we performed LC-MS analysis of the inactivated protein. For these studies we incubated rabbit 2E1 with DDC in the reconstituted system. These studies were performed with rabbit 2E1 because it can readily be separated from reductase, whereas we have not yet been able to separate human CYP2E1 from reductase. As shown in Fig. 4A, the native unmodified CYP2E1 exhibited a single peak with a mass of 53,798 Da in the MS spectrum. This value corresponds well with the molecular mass predicted on the basis of the amino acid sequence as well as previously published results (Blobaum et al., 2002). After incubation of CYP2E1 with DDC and NADPH for 30 min, two mass peaks were observed in the MS spectrum at 53,800 and 53,915 Da (Fig. 4B). The former mass is that for the unmodified CYP2E1 and the latter is that for the modified CYP2E1, which exhibited an increase in mass of approximately 115 Da.
Fig. 4.
Deconvoluted mass spectra for the native CYP2E1 (A) and the inactivated CYP2E1 (B) after reaction with diethyldithocarbamate under turnover conditions. The inactivations were performed as described under Materials and Methods. Aliquots of the primary reaction mixture containing 50 pmol of CYP2E1 were loaded onto a C4 column, and the CYP2E1 samples were separated from the rest of the components in the reconstituted mixture as described under Materials and Methods. The molecular masses of the CYP2E1 samples were analyzed by ESI-LC/MS as described under Materials and Methods.
The magnitude of this change in the mass of the protein suggested that DDC might be forming a disulfide bond with a cysteine on CYP2E1. To test for the presence of a disulfide bond in the protein that could be cleaved by the addition of DTT, we performed the inactivation by DDC as before and then we performed a second incubation for 30 min at 30°C in the presence of 10 mM DTT. We then analyzed the samples by ESI-LC/MS and found that the sample incubated with DTT had lost the adduct and exhibited a mass spectrum similar to that in Fig. 4A, whereas a sample incubated in the absence of DTT retained the adduct. However, the activity of the protein could not be regained after incubation in the presence of DTT (data not shown).
LC-MS/MS Analysis of a Reactive Metabolite of DDC Formed by CYP2E1.
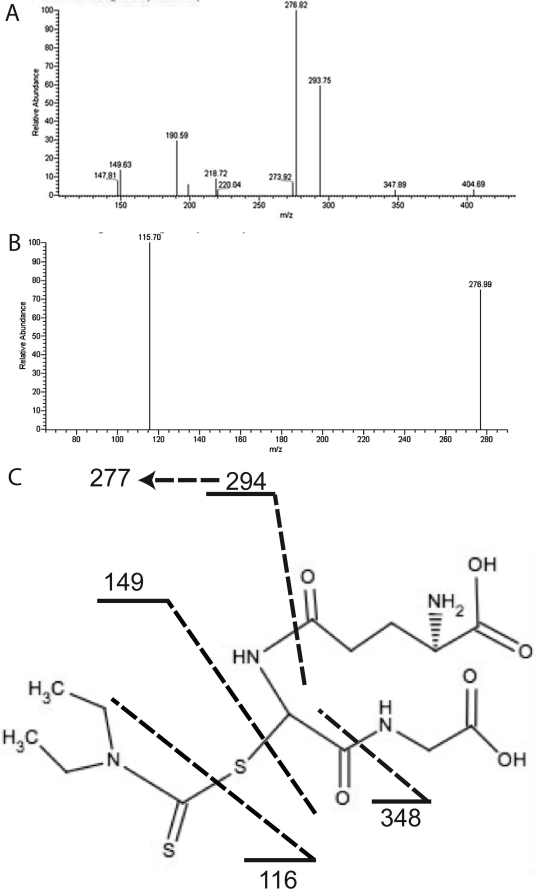
The formation of reactive intermediates during the metabolism of DDC by CYP2E1 in the reconstituted system was investigated by trapping the intermediate(s) with GSH and analyzing the product by LC-MS/MS. LC-MS/MS analysis of the reaction mixture indicated the formation of one metabolite that eluted at approximately 14.3 min with the MH+ at m/z 423. The proposed structure (Fig. 5C) of the adduct is based on the fragmentation patterns shown in Fig. 5, A and B. In Fig. 5A, the fragment at m/z 404 in the MS2 spectrum of the GSH conjugate is due to the loss of oxygen. The fragment at 347 is due to neutral loss of glycine (75 Da). The fragment at m/z 293 is the product of neutral loss of anhydroglutamic acid (129 Da). The fragment at m/z 277 is the product of neutral loss of glutamine (146 Da). The fragments at m/z 218 and 190 are the products from the loss of both glycine and anhydroglutamic acid and then the subsequent loss of carbon monoxide. The peak at 149 is the S-(N,N-diethylthiocarbamoyl). The peak at 115 in the MS3 spectrum of the 276 fragment (Fig. 5B) is due to the additional loss of the sulfur from the S-(N,N-diethylthiocarbamoyl). The spectrum in Fig. 5A is almost identical to that reported by Jin et al. (1994) for S-(N,N-diethylthiocarbamoyl)glutathione.
Fig. 5.
LC-MS/MS analysis of the DDC-GSH conjugate formed during metabolism by reconstituted CYP2E1. A, MS2 spectrum of the DDC-GSH conjugate eluting at 14.34 min with an m/z of 423. B, MS3 spectrum from the 276 fragment. C, the proposed structure of the DDC-GSH conjugate and the dashed lines are the sites of fragmentation as described in the text.
Discussion
The use of disulfiram in studies aimed to inactivate CYP2E1 in vivo has been reported recently (Doroshyenko et al., 2009); however, the mechanism by which it causes inactivation has not been reported. We report here that disulfiram is not the compound directly responsible for inactivation of CYP2E1, but that its reduced form, diethyldithiocarbamate, is responsible for the inactivation (Fig. 1). Using purified human CYP2E1, we were able to show that disulfiram does not inactivate P450 at concentrations up to 20 mM; however, DDC caused a significant loss in activity in a time- and concentration-dependent manner in the presence of NADPH. These results indicate that for studies performed in vivo disulfiram must first be reduced in the body to DDC before its interaction with CYP2E1, leading to inactivation.
We have also characterized the kinetics for the inactivation of wild-type CYP2E1 as well as two of its single amino acid genetic polymorphic forms. The wild-type 2E1 exhibits a KI of 12.2 μM and a kinact of 0.020 min−1 (Table 1). The polymorphic CYP2E1.4 demonstrated characteristics similar to those of the wild type with a KI of 12.5 μM and a kinact of 0.0187 min−1. This result was not surprising because previous studies have also shown that this mutation has no affect on enzymatic activity (Hu et al., 1997). The polymorphic CYP2E1.2 had previously been reported to exhibit slower rates of metabolism than the wild type for some substrates (Hu et al., 1997), as well as faster rates for others (Hanioka et al., 2003). We found that DDC is not a very good inactivator of CYP2E.2, exhibiting a KI of 227.6 μM and a kinact of 0.0061 min−1 (Table 1). Thus, in terms of overall efficiency, as measured by kinact/KI, the inactivation of CYP2E1.2 is 1000-fold less efficient than the inactivation of the wild-type 2E1. The CYP2E1.2 polymorphism is a change of residue 76, which is conserved in many mammalian species, from an arginine to a histidine (Song et al., 1986; Khani et al., 1988; Freeman et al., 1992; Komori et al., 1992). The crystal structure of CYP2E1 indicates that Arg-76 is part of the β1–2 structural unit, but that it is oriented toward the solvent (Porubsky et al., 2008). Therefore, it is difficult to explain the significant impact of the mutation on the catalytic functionality of CYP2E1; however, possible explanations could include alterations in the substrate entry or product egress channels, effects on the incorporation of the 2E1 into the membrane, or effects on the protein dynamics of the P450. The lack of any significant alteration in the catalytic properties of the polymorphism in CYP2E1.4, in which an isoleucine is substituted for a valine, is probably due to the structural similarities of the two amino acid residues and the fact that position 179 is on the periphery of the protein.
Diode array spectral analysis of the metabolite of DDC formed by 2E1 indicates a change in the UV spectrum from having two peaks at 244 and 323 to a single peak at 221, and the metabolite exhibits a change in elution time on HPLC from 29.5 to 27.7 min (Fig. 3). No other peaks were observed in the elution profile for the metabolism of DDC by CYP2E1, which indicates that either this product was the only one produced by 2E1 or that other products could not be detected spectrally.
To determine the mass of the apoprotein adduct formed by the metabolism of DDC to a reactive intermediate, we incubated rabbit CYP2E1 with DDC and then analyzed the modified protein by electrospray ionization liquid chromatography-mass spectrometry. Our results demonstrated that the loss in the catalytic activity of CYP2E1 is due to the formation of a single adduct of DDC with the protein. The evidence for this observation comes from the increase in the observed mass of the inactivated CYP2E1 apoprotein of 115 mass units (Fig. 4B). In addition, HPLC analysis of the inactivated protein reaction mixture showed no change in the unmodified heme content or the presence of any modified heme (data not shown).
To determine a more accurate mass of the adduct, we performed a GSH trapping experiment. Data from the CYP2E1 protein adduct as well as products observed by Jin et al. (1994) helped to narrow down the mass range to search. Jin et al. (1994) administered disulfiram to mice and used mass spectrometry to examine GSH conjugates in the bile. They observed five DDC metabolites and the second most prevalent product, S-(N,N-diethylthiocarbamoyl)glutathione, exhibited an MH+ ion at m/z 423. This would correspond to a DDC metabolite of 116 Da, which is within the error for the mass of the adduct we observed with the apoprotein. Analysis of the products formed after incubation of DDC with reconstituted human CYP2E1 and GSH indicated the formation of a GSH adduct with a MH+ ion at m/z 423. Analysis of the fragmentation pattern indicated peaks that corresponded to the characteristic loss of glycine, anhydroglutamic acid, and glutamine.
We postulate that this metabolite could inactivate CYP2E1 by forming a disulfide bond with one of the eight cysteines that are present in the apoprotein. The loss of the DDC adduct and reversion of the mass of the 2E1 to that of the unmodified protein in the presence of DTT provide addition evidence in support of this hypothesis. We are currently attempting to determine which of the eight cysteine residues in CYP2E1 is covalently modified by the metabolite of DDC, leading to the inactivation. Attempts to digest the modified protein, separate it by LC, and sequence it by MS/MS have so far not been successful, presumably because of the lability of the disulfide bond. Studies involving site-specific mutagenesis of the cysteines are in progress.
This work was supported in part by the National Institutes of Health National Cancer Institute [Grant CA16954] (to P.F.H.); and the National Institutes of Health National Institute on Drug Abuse [Biology of Drug Abuse Postdoctoral Training Fellowship DA007268-18] (to M.P.-H.).
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.034710.
- ALDH
- aldehyde dehydrogenase
- DBH
- dopamine-β-hydroxylase
- DDC
- diethyldithiocarbamate
- P450
- cytochrome P450
- 7-EFC
- 7-ethoxy-4-(trifluoromethyl)-coumarin
- HPLC
- high-performance liquid chromatography
- ESI
- electrospray ionization
- LC
- liquid chromatography
- MS
- mass spectrometry
- MS/MS
- tandem mass spectrometry
- DTT
- dithiothreitol.
References
- Blobaum AL, Kent UM, Alworth WL, Hollenberg PF. (2002) Mechanism-based inactivation of cytochromes P450 2E1 and 2E1 T303A by tert-butyl acetylenes: characterization of reactive intermediate adducts to the heme and apoprotein. Chem Res Toxicol 15:1561–1571 [DOI] [PubMed] [Google Scholar]
- Cobby J, Mayersohn M, Selliah S. (1977) The rapid reduction of disulfiram in blood and plasma. J Pharmacol Exp Ther 202:724–731 [PubMed] [Google Scholar]
- Doroshyenko O, Fuhr U, Kunz D, Frank D, Kinzig M, Jetter A, Reith Y, Lazar A, Taubert D, Kirchheiner J, et al. (2009) In vivo role of cytochrome P450 2E1 and glutathione-S-transferase activity for acrylamide toxicokinetics in humans. Cancer Epidemiol Biomarkers Prev 18:433–443 [DOI] [PubMed] [Google Scholar]
- Fairbrother KS, Grove J, de Waziers I, Steimel DT, Day CP, Crespi CL, Daly AK. (1998) Detection and characterization of novel polymorphisms in the CYP2E1 gene. Pharmacogenetics 8:543–552 [DOI] [PubMed] [Google Scholar]
- Freeman JE, Stirling D, Russell AL, Wolf CR. (1992) cDNA sequence, deduced amino acid sequence, predicted gene structure and chemical regulation of mouse Cyp2e1. Biochem J 281 (Pt 3):689–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaval-Cruz M, Weinshenker D. (2009) Mechanisms of disulfiram-induced cocaine abstinence: antabuse and cocaine relapse. Mol Interv 9:175–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein M. (1966) Inhibition of norepinephrine biosynthesis at the dopamine-beta-hydroxylation stage. Pharmacol Rev 18:77–82 [PubMed] [Google Scholar]
- Guengerich FP, Kim DH, Iwasaki M. (1991) Role of human cytochrome P-450 IIE1 in the oxidation of many low molecular weight cancer suspects. Chem Res Toxicol 4:168–179 [DOI] [PubMed] [Google Scholar]
- Hald J, Jacobsen E. (1948) A drug sensitizing the organism to ethyl alcohol. Lancet 2:1001–1004 [DOI] [PubMed] [Google Scholar]
- Hanioka N, Tanaka-Kagawa T, Miyata Y, Matsushima E, Makino Y, Ohno A, Yoda R, Jinno H, Ando M. (2003) Functional characterization of three human cytochrome p450 2E1 variants with amino acid substitutions. Xenobiotica 33:575–586 [DOI] [PubMed] [Google Scholar]
- Hu Y, Oscarson M, Johansson I, Yue QY, Dahl ML, Tabone M, Arincò S, Albano E, Ingelman-Sundberg M. (1997) Genetic polymorphism of human CYP2E1: characterization of two variant alleles. Mol Pharmacol 51:370–376 [PubMed] [Google Scholar]
- Jin L, Davis MR, Hu P, Baillie TA. (1994) Identification of novel glutathione conjugates of disulfiram and diethyldithiocarbamate in rat bile by liquid chromatography-tandem mass spectrometry. Evidence for metabolic activation of disulfiram in vivo. Chem Res Toxicol 7:526–533 [DOI] [PubMed] [Google Scholar]
- Kent UM, Roberts ES, Chun J, Hodge K, Juncaj J, Hollenberg PF. (1998) Inactivation of cytochrome P450 2E1 by tert-butylisothiocyanate. Chem Res Toxicol 11:1154–1161 [DOI] [PubMed] [Google Scholar]
- Khani SC, Porter TD, Fujita VS, Coon MJ. (1988) Organization and differential expression of two highly similar genes in the rabbit alcohol-inducible cytochrome P-450 subfamily. J Biol Chem 263:7170–7175 [PubMed] [Google Scholar]
- Kharasch ED, Hankins DC, Jubert C, Thummel KE, Taraday JK. (1999) Lack of single-dose disulfiram effects on cytochrome P-450 2C9, 2C19, 2D6, and 3A4 activities: evidence for specificity toward P-450 2E1. Drug Metab Dispos 27:717–723 [PubMed] [Google Scholar]
- Kitson TM. (1977) The disulfiram-ethanol reaction: a review. J Stud Alcohol 38:96–113 [DOI] [PubMed] [Google Scholar]
- Komori M, Kikuchi O, Sakuma T, Funaki J, Kitada M, Kamataki T. (1992) Molecular cloning of monkey liver cytochrome P-450 cDNAs: similarity of the primary sequences to human cytochromes P-450. Biochim Biophys Acta 1171:141–146 [DOI] [PubMed] [Google Scholar]
- Koop DR. (1992) Oxidative and reductive metabolism by cytochrome P450 2E1. FASEB J 6:724–730 [DOI] [PubMed] [Google Scholar]
- Larson JR, Coon MJ, Porter TD. (1991) Alcohol-inducible cytochrome P-450IIE1 lacking the hydrophobic NH2-terminal segment retains catalytic activity and is membrane-bound when expressed in Escherichia coli. J Biol Chem 266:7321–7324 [PubMed] [Google Scholar]
- Lieber CS. (1997) Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev 77:517–544 [DOI] [PubMed] [Google Scholar]
- Lin HL, Kent UM, Hollenberg PF. (2005) The grapefruit juice effect is not limited to cytochrome P450 (P450) 3A4: evidence for bergamottin-dependent inactivation, heme destruction, and covalent binding to protein in P450s 2B6 and 3A5. J Pharmacol Exp Ther 313:154–164 [DOI] [PubMed] [Google Scholar]
- Lin HL, Zhang H, Hollenberg PF. (2009) Metabolic activation of mifepristone [RU486; 17β-hydroxy-11β-(4-dimethylaminophenyl)-17α-(1-propynyl)-estra-4,9-dien-3-one] by mammalian cytochromes P450 and the mechanism-based inactivation of human CYP2B6. J Pharmacol Exp Ther 329:26–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsky JJ, Shen ML, Naylor S. (2001) In vivo inhibition of aldehyde dehydrogenase by disulfiram. Chem Biol Interact 130–132:93–102 [DOI] [PubMed] [Google Scholar]
- Musacchio JM, Goldstein M, Anagnoste B, Poch G, Kopin IJ. (1966) Inhibition of dopamine-β-hydroxylase by disulfiram in vivo. J Pharmacol Exp Ther 152:56–61 [PubMed] [Google Scholar]
- Ploemen JP, van Iersel ML, Wormhoudt LW, Commandeur JN, Vermeulen NP, van Bladeren PJ. (1996) In vitro inhibition of rat and human glutathione S-transferase isoenzymes by disulfiram and diethyldithiocarbamate. Biochem Pharmacol 52:197–204 [DOI] [PubMed] [Google Scholar]
- Porubsky PR, Meneely KM, Scott EE. (2008) Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J Biol Chem 283:33698–33707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EE, Spatzenegger M, Halpert JR. (2001) A truncation of 2B subfamily cytochromes P450 yields increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys 395:57–68 [DOI] [PubMed] [Google Scholar]
- Song BJ, Gelboin HV, Park SS, Yang CS, Gonzalez FJ. (1986) Complementary DNA and protein sequences of ethanol-inducible rat and human cytochrome P-450s. Transcriptional and post-transcriptional regulation of the rat enzyme. J Biol Chem 261:16689–16697 [PubMed] [Google Scholar]
- Wattenberg LW, Sparnins VL, Barany G. (1989) Inhibition of N-nitrosodiethylamine carcinogenesis in mice by naturally occurring organosulfur compounds and monoterpenes. Cancer Res 49:2689–2692 [PubMed] [Google Scholar]
- Zhang H, Lin HL, Walker VJ, Hamdane D, Hollenberg PF. (2009) tert-Butylphenylacetylene is a potent mechanism-based inactivator of cytochrome P450 2B4: inhibition of cytochrome P450 catalysis by steric hindrance. Mol Pharmacol 76:1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]