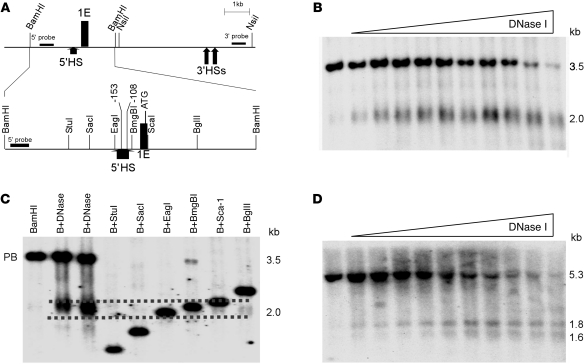
Figure 1. Mapping DNase I HSs in the erythroid promoter region of the ankyrin-1 locus in erythroid cells.
(A) The map indicates the position of ankyrin exon 1E, flanking restriction enzyme sites, and probes used in DNase I mapping. A close-up view of the BamHI fragment with appropriate restriction enzyme sites, the –108 and –153 mutations, and exon 1E is also shown. Corresponding coordinates are (GRCh37/hg19) BamHI 41657070; StuI 41656031; SacI 41655737; EagI 41655310; 153 mutation 41655209; –108 mutation 41655164; BmgBI 41655152; ATG. (B) Mapping 5′ of exon 1E. Nuclei from K562 cells were treated with increasing amounts of DNase I, digested with BamHI, and subjected to Southern blot analysis using the 5′ probe in A. This yielded the expected 3.5-kb BamHI fragment and a smaller, 2.0-kb fragment corresponding to a DNase I HS. (C) Fine mapping the 5′ HS. Human genomic DNA digested with BamHI (B) or with BamHI and the indicated second enzyme. DNA from DNase I–treated K562 nuclei was also digested with BamHI (B+DNase). These digests were subjected to Southern blot analysis using the 5′ probe in A. Dashed lines indicate the estimated upper and lower limits of 5′HS. The undigested parent band is indicated (PB). (D) Mapping 3′ of exon 1E. Nuclei from K562 cells were treated with increasing amounts of DNase I, digested with NsiI, and subjected to Southern blot analysis using the 3′ probe in A. This yielded the expected 5.3-kb NsiI fragment and two smaller, 1.8-kb and 1.6-kb fragments, corresponding to DNase I HSs.