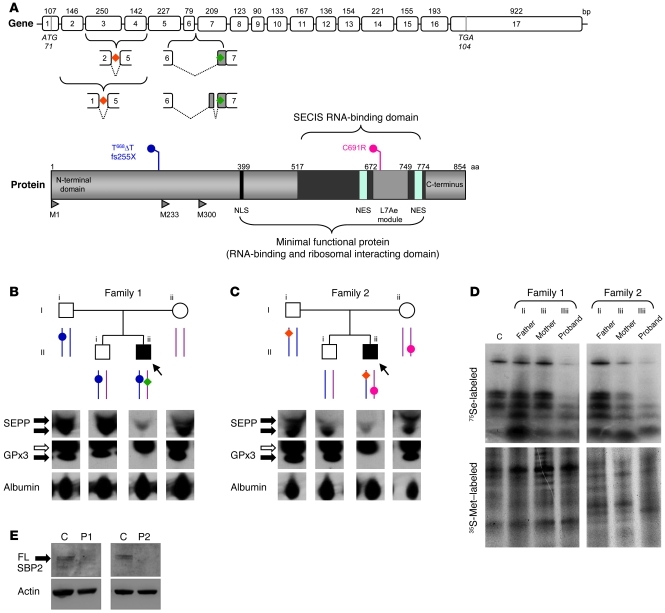
Figure 1. Genotypes and biochemical phenotypes in 2 families with defects in SECISBP2.
(A) Schematic representations of the coding exons of SECISBP2 (top) showing the location of aberrantly spliced variant transcripts (P1, green; P2, orange), and the major domains of SBP2 (bottom) showing the frameshift premature stop (P1, blue, fs255X) and missense (P2, red, C691R) mutations. NLS, nuclear localization signal; NES, nuclear export signal; L7Ae module, domain homolog to RNA-binding domain of L7Ae; M1, M233, M300, position of alternate methionine transcription initiation sites, with M1 yielding full-length wild-type SBP2 protein. (B and C) Pedigrees of families 1 (B) and 2 (C) showing genotypes (as described in A); squares and circles represent male and female family members, respectively, and arrows/filled symbols denote probands. Below, Western blotting for SEPP and GPx3 in plasma of both probands and family members. Black arrows indicate specific bands; white arrows denote nonspecific bands. Albumin was used as a loading control. (D) PBMCs from P1 and P2 demonstrate defective selenoprotein synthesis. Selenoprotein biosynthesis in PBMCs was assessed using 75Se labeling as described in Methods. 35S-Met labeling confirmed comparable protein loading. C, control subject (age- and sex-matched to P1). (E) Fibroblasts from P1 and P2 lack full-length SBP2. Whole cell lysates from primary skin fibroblasts were Western blotted for full-length (FL) SBP2 (arrow) with actin as a loading control. Age- and sex-matched control subjects were used.