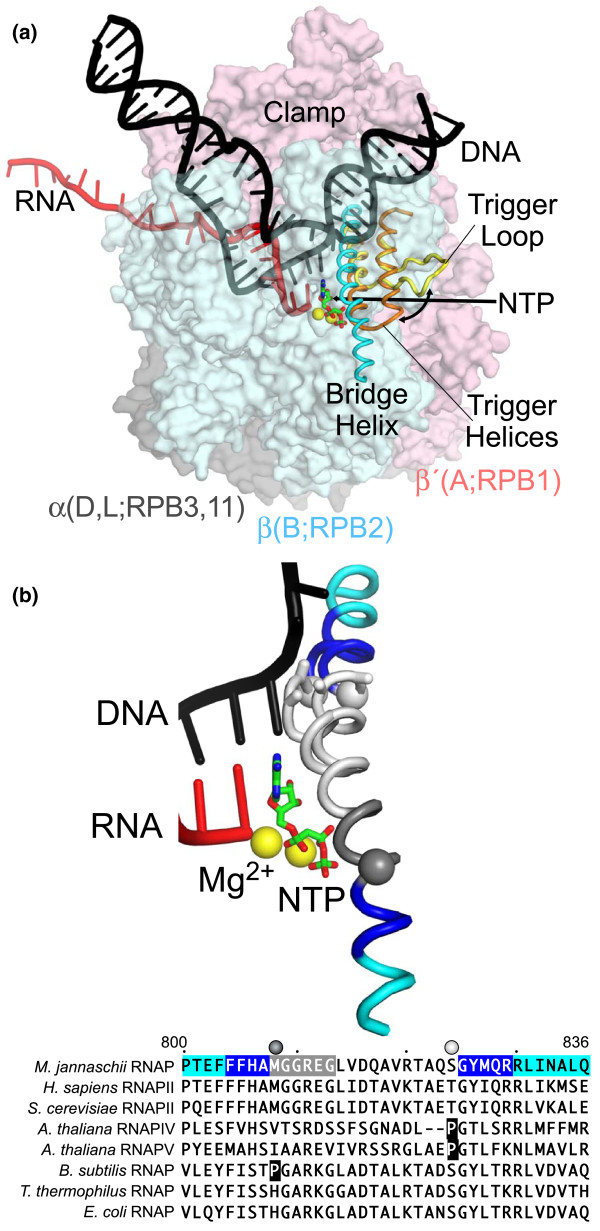
Figure 1.
Positions and conformations of the bridge helix in an elongation complex. (a) Structure of an elongation complex based on the crystal structure of a NTP-bound RNA polymerase from Thermus thermophilus (PDB 2o5j) [3]. DNA (black) is melting into a transcription bubble that allows template-strand pairing with RNA (red) in a 9-10 base pair RNA-DNA hybrid. The bridge helix (cyan) and trigger loop/helices (yellow/orange) lie on the downstream side of the active site. The presumed path of NTP entry is indicated by the straight arrow. Interconversion of the trigger loop and trigger helices is indicated by the curved arrow. The RNA polymerase subunits are shown as semi-transparent surfaces with the identities of orthologous subunits in bacteria (α, β, and β', gray, blue, and pink, respectively), archaea (D, L, B, and A), and eukaryotic RNA polymerase II (RPB3, 11, RPB2, RPB1) indicated. The active site Mg2+ ions are shown as yellow spheres, and α,β-methylene-ATP in green and red. (b) Conformations of the bridge helix observed on crystal structures of a NTP-bound elongation complex and of an RNA polymerase lacking nucleic acids. The positions of nascent RNA, the template DNA strand, α,β-methylene-ATP, Mg2+, and straight bridge helix are from the PDB 2o5j structure. The looped-out bridge helix indicating the conformation in a nucleic-acid-free structure is from T. thermophilus RNA polymerase bound by σA initiation factor (PDB 1iw7). Positions at which substitutions with proline increase polymerase activity are marked by Cα spheres (HN and HC) [7]. The location of a deletion of two amino acids in the plant RNA polymerase IV enzyme is marked by Cα-Cβ sticks (next to the white sphere marking the proline substitution). Sequences of the bridge helix from several RNA polymerases are shown, with the M. jannaschii bridge helix color-coded as in the molecular model: blue, segments in which two-amino-acid deletions eliminate polymerase activity; gray, segment in which deletions partially affect activity; white, segment in which deletions have minimal effect on activity; cyan, amino- and carboxy-terminal segments. Naturally occurring prolines at HN and HC are shown white-on-black.