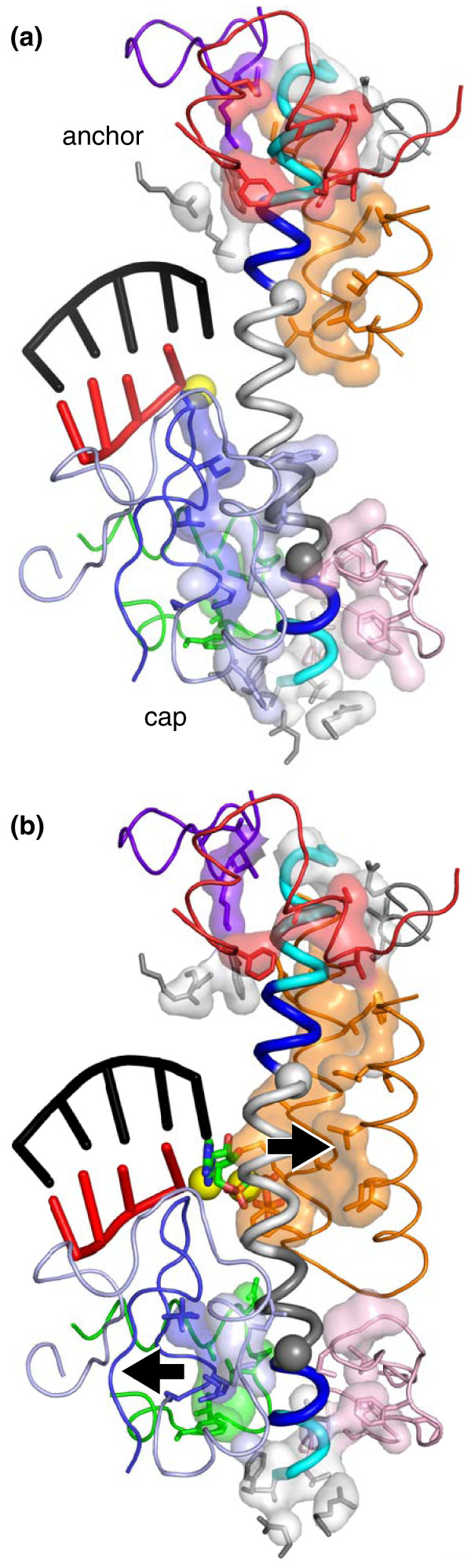
Figure 2.
RNA polymerase residues that contact the bridge helix. DNA downstream of the active site is omitted for clarity. (a) Contacts in a T. thermophilus elongation complex lacking NTP (PDB 2o5i). (b) Contacts in a T. thermophilus elongation complex bound by α,β-methylene-ATP (PDB 2o5j). Residues that lie within 4 Å of the bridge helix (contacts) are shown as a semi-transparent surface and as sticks. Contacts occur principally in two regions, a cap that contacts the amino-terminal portion of the bridge helix and, in the NTP-bound complex, the trigger helices. Contacts made by polymerase loops or modules that change upon bridge-helix movements are color-coded: blue, RPB2/β D-loop; light blue, RPB2/β fork loop; green, RPB2/β link loop and helix; light pink, RPB1/β' F-loop; red, RPB1/β' switch 1; purple, RPB1/β' switch 5 and 11 adjacent residues; orange, RPB1/β' trigger loop or trigger helices. A portion of the trigger loop in the NTP-free elongation complex that does not contact the bridge helix is not shown and was not ordered in the structure. Other segments or individual side chains contacting the bridge helix are shown but not colored. Arrows indicate small movements of the bridge helix, D-loop, and fork loop (all approximately 1.5 Å) that occur upon substrate binding coupled to a larger movement of the RPB2/β lobe [3].