Figure 1.
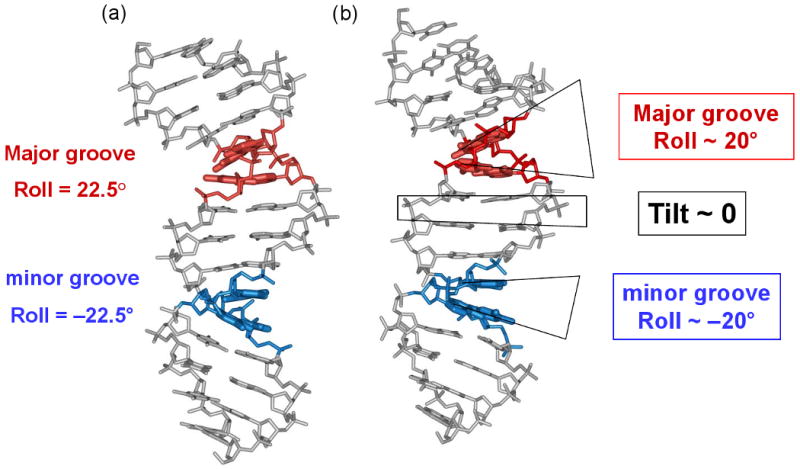
Comparison of the “mini-kink” model for nucleosomal DNA bending (22, 38), (a), and the crystallographic structure of a nucleosome (54), (b). (a) According to the model, bends into the major and minor grooves occur predominantly at the YR (red) and RY (blue) dimers, respectively. (b) Directionality and extent of the DNA bending in the nucleosomal X-ray structure (54) are generally consistent with the earlier model shown in (a). The DNA fragment shown corresponds to the superhelical locations SHL −4 and −3.5 in the X-ray structure 1KX3 (54). The color coding is the same as in (a): red for positive Roll (bending into the major groove) and blue for negative Roll (bending into the minor groove). The actual Roll values are about ±15-20°, which is close to ±22.5° estimated by the “mini-kink” model (22, 38).
