Figure 1.
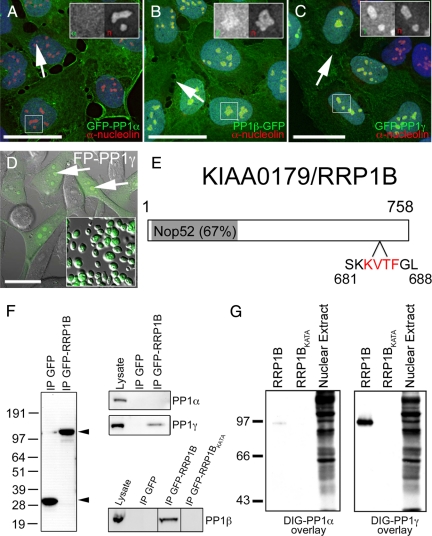
Identification of RRP1B as a nucleolar targeting subunit that specifically binds PP1β and PP1γ. (A) When stably expressed at low levels in U2OS cells as a GFP fusion protein, PP1α (green) is found distributed throughout the cell. (B) Stably expressed GFP-tagged PP1β (green) is also found throughout the cell but shows an additional accumulation in nucleoli (arrows), as does stably expressed GFP-PP1γ (C, green). DNA stained with Hoechst 33342 is shown in blue and anti-nucleolin, a nucleolar marker protein, in red. For each panel there is an inset showing an expanded version of the boxed region with the GFP-PP1 signal on the left and the anti-nucleolin signal on the right. (D) GFP-PP1γ (green), shown here superimposed on a differential interference contrast (DIC) image in HeLaGFP-PP1γ cells, accumulates in nucleoli (arrows) and retains its nucleolar association in purified nucleoli (inset). Scale bars are 15 μm. (E) RRP1B is a 758-aa protein that contains a Nop52 domain (67% identical to yeast NOP52) and a putative PP1 binding RVxF motif (aa 683-686). (F) GFP-Trap pulls down similar amounts of free GFP and GFP-RRP1B from U2OSGFP and U2OSGFP-RRP1B cells, respectively. Endogenous PP1γ, but not PP1α, copurifies specifically with GFP-RRP1B. Endogenous PP1β also copurifies with GFP-RRP1B, but not with the GFP-RRP1BKATA mutant in which the PP1 binding domain has been disrupted. (G) Far Western blot demonstrating that RRP1B preferentially binds PP1γ over PP1α and that the KATA mutation greatly weaken its ability to bind PP1. Equal amounts of purified recombinant DIG-labeled PP1α and PP1γ were overlayed on 200 ng of purified recombinant wild type and KATA mutant RRP1B. The control lane (10 μg rat liver nuclear extract) illustrates that DIG-PP1α binds other target proteins with high affinity.