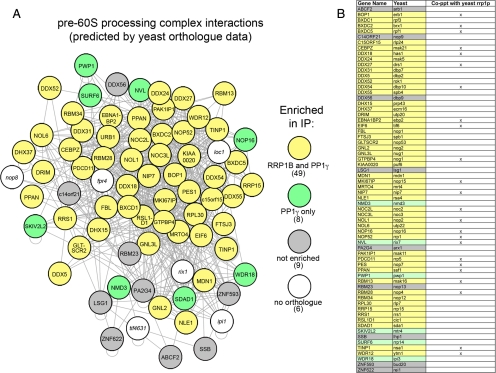
Figure 8.
RRP1B and PP1γ copurify an overlapping subset of pre-60S ribosomal subunit processing proteins. (A) An overlay of mammalian orthologues on their yeast counterparts in a yeast-derived map of pre-60S ribosomal subunit processing proteins demonstrates a specific enrichment of midlate processing complexes. Of the 72 proteins predicted in the yeast map, 66 have known mammalian orthologues. Of these, 49 (74%) were found in both the RRP1B and PP1γ interactomes (yellow), with an additional eight proteins only detected with the phosphatase (green). This may represent different subcomplexes or a difference in sensitivity of the experiment. Of the nine proteins that were not enriched (gray), most are peripheral to the core complex and function later in the processing/export pathway. In addition to these nonribosomal processing proteins, 36/40 (90%) of 60S subunit RPL proteins were specifically enriched with RRP1B and PP1γ. (B) 60S processing proteins identified in a recent nonquantitative screen of TAP-tagged yeast Rrp1p are indicated in this table, which includes both the yeast and mammalian gene names for each protein and uses the same color coding as in A to indicate whether a protein was found in our RRP1B and/or PP1γ IP or neither.