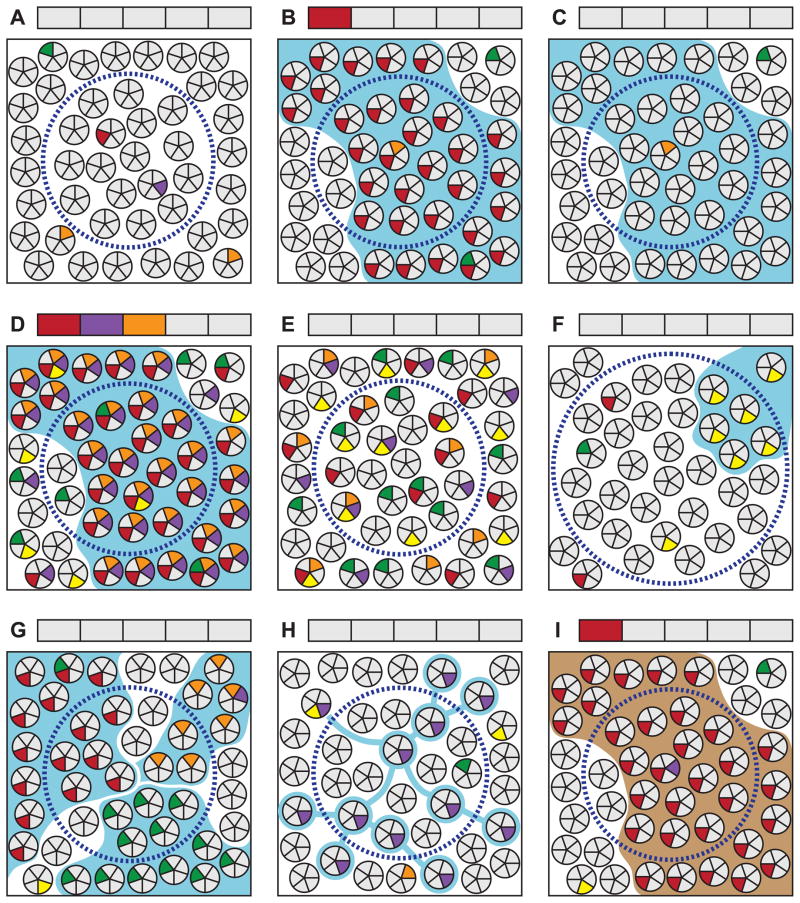
Figure 3.
Selected examples of important factors for consideration when interpreting clonality studies. See section 9 in the text for a detailed discussion of each scenario. Clusters of small circles in each panel represent a population of cells in a tissue. Each wedge represents a genomic site; gray and colored wedges respectively indicate wild-type and mutant genotypes. Blue boundaries indicate the true clonal relationship of cells derived from a neoplastic clonal expansion. Brown boundaries indicate an embryonically established clonal relationship. Dashed circles represent an area sampled for analysis. Boxes above each panel correspond to the genomic sites in the cell wedges and indicate the sample genotype as reported by a conventional method of sequencing that requires the majority of harvested cells to bear a mutation for it to be detectable. (A) Random mutations arising within individual cells in a tissue are not detectable by standard techniques. (B) If a cell bearing a unique mutation undergoes clonal expansion, it will become detectable when assessing this site. Note that new random mutations arise in some cells during clonal expansion which are not detected. (C) Clonal expansions will not be detectable if none of the genomic sites screened happen to be uniquely mutated in the founder cell. The probability of detecting a clone is a function of the number of sites screened and the frequency of random mutations in the founder cell. The latter, in turn, is dependent upon the per-cell-division mutation rate of each site and the number of cell divisions having occurred between the zygote and clone’s founding. (D) An elevated mutation frequency will facilitate a clone’s detection, although (E) in the absence of clonal expansion will have no effect on the detectable mutation load. (F) Clones that are small relative to the population sampled or that (G) become extensively mixed with other clones or (H) adjacent normal cells during expansion, are not detectable by conventional sequencing methods. (I) Based only on the presence of a detectable mutation, clonal patches arising during embryogenesis will be indistinguishable from those derived from neoplastic processes. Mutation spectrum, frequency and other contextual factors must be relied upon to infer the distinction.