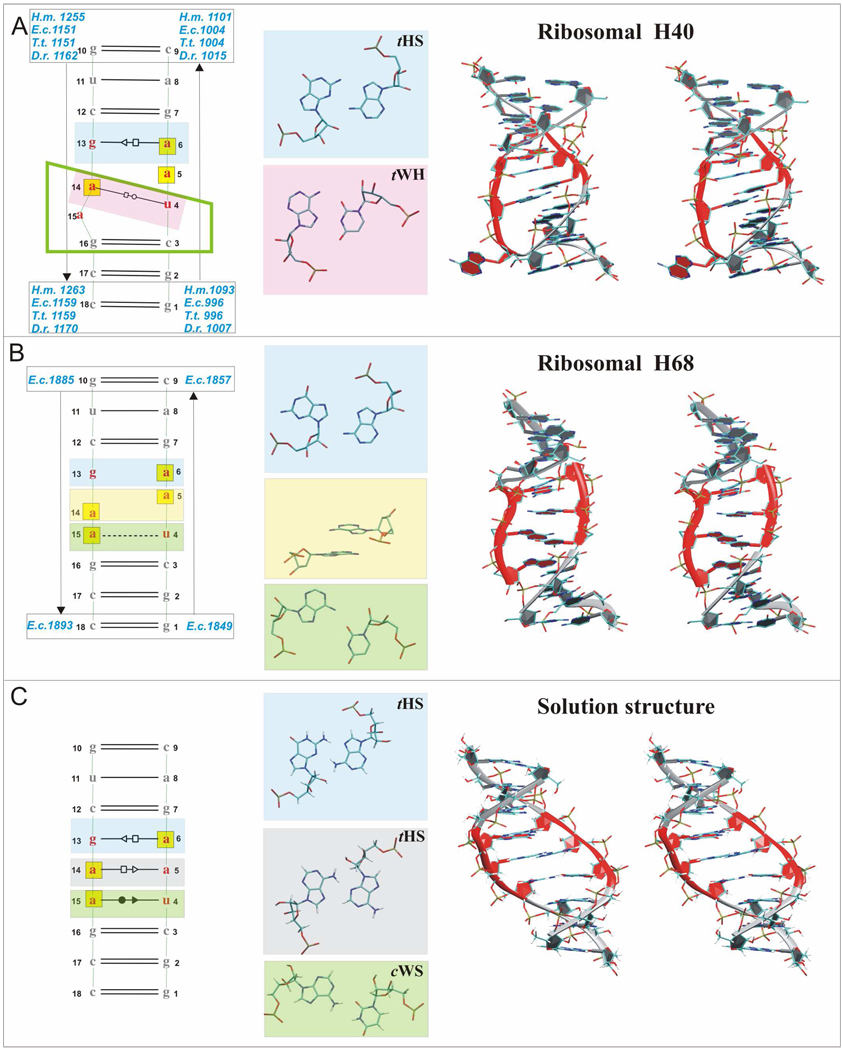
Figure 1.
2D structures (Left) and 3D stereo views (Right) of studied segments including non-Watson-Cricks base pairs in the internal UAA/GAA loop (Middle). A. Left: 2D X-ray structure of H40 with unified sequence flanking the internal loop (see Materials and Methods). Middle: sheared A/G and rH U/A base pairs. Right: Stereo view of E.c. H40 X-ray structure. B. Left: 2D X-ray structure of E.c. 23S rRNA H68 exhibiting an alternative conformation of UAA/GAA motif with unified canonical flanking sequence. Black dashed line indicates single H-bond. Middle: unpaired G and A bases, stacking middle adenines and single bonded A/U base pair. Right: Stereo view of this structure. C. Left: 2D NMR structure. Middle: sheared A/G, sheared A/A and incomplete cWS A/U base pairs. Right: Stereo view of the NMR structure. In all Figures, bases of the UAA/GAA internal loop are in red, 3D structures are colored accordingly, hydrogens are not shown in the X-ray structures, bases in yellow boxes in the 2D structures are involved in stacking and the marks between the bases indicate base paring family according to the Leontis & Westhof classification (tHS = trans Hoogsteen/Sugar edge A/G or A/A, known also as “sheared” base pairs, tWH = trans Watson-Crick/Hoogsteen U/A, known also as reverse Hoogsteen (rH) base pair, and cWS = cis Watson-Crick/Sugar edge A/U).6,7 X-ray nucleotide numbers are in blue, NMR numbers are in black. The green rectangular trapezium for H40 structure marks bases forming the UA_handle submotif.8