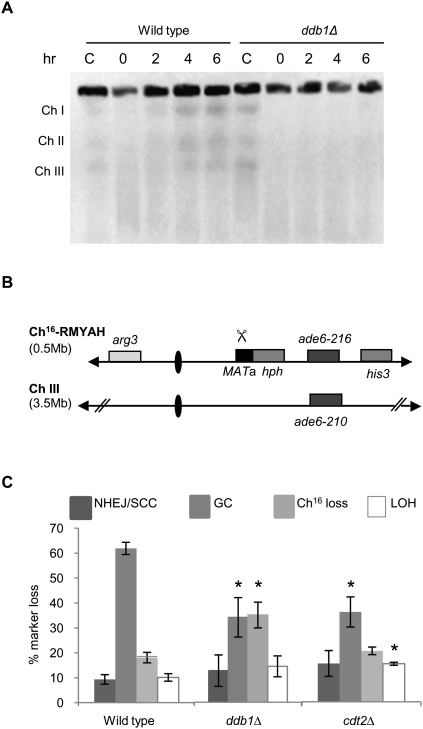
Figure 2.
Ddb1 and Cdt2 are required for efficient HR repair. (A) PFGE analyses of chromosomes (Ch) from bleomycin-treated (5 μg/mL) wild-type and ddb1Δ cells. Cells were either untreated (C) or treated with bleomycin for 1 h, washed to remove bleomycin, and harvested at the times indicated following bleomycin removal. (B) Schematic of the minichromosome Ch16-RMYAH. Ch16-RMYAH, a 530-kb fragment of ChIII, contains the MATa site with an adjacent hygromycin resistance marker gene (hph) and the ade6-M216 ade heteroallele, complemented by ade6-M210, present on ChIII, in addition to a his3 and arg3 markers, as shown previously (Tinline-Purvis et al. 2009). Derepression of pREP81X-HO (not shown) generates a DSB at the MATa target site (indicated by scissors). (C) Percentage of DSB-induced marker loss in wild-type, ddb1Δ, and cdt2Δ backgrounds. Means ± standard errors of three experiments are shown. The asterisk (*) represents significant difference compared with wild type.