Abstract
Surface-bound nucleic acid probes designed to adopt specific secondary structures are becoming increasingly important in a range of biosensing applications but remain less well characterized than traditional single-stranded probes, which are typically designed to avoid secondary structure. We report the hybridization efficiency for surface-immobilized hairpin DNA probes. Our probes are molecular beacons, carrying a 3′ dye moiety and a 5′ thiol for attachment to gold nanowires, which serve as both scaffolds for probe attachment and quenchers. Hybridization efficiency was dependent on probe surface coverage, reaching a maximum of ∼90% at intermediate coverages of (1−2) × 1012 probes/cm2 and dropping to ≤20% at higher or lower coverages. Fluorescence intensity did not track with the number of target molecules bound, and was highest for high probe coverage despite the lower bound targets per square centimeter. Backfilling with short thiolated oligoethylene glycol spacers increased hybridization efficiency at low hairpin probe coverages (∼(3−4) × 1011 probes/cm2), but not at higher probe coverages (1 × 1012/cm2). We also evaluated the effect of target length by adding up to 50 nonhybridizing nucleotides to the 3′ or 5′ end of the complementary target sequence. Additional nucleotides on the 3′ end of the complementary target sequence (i.e., the end near the nanowire surface) had a much greater impact on hybridization efficiency as compared to nucleotides added to the 5′ end. This work provides guidance in designing sensors in which surface-bound probes designed to adopt secondary structures are used to detect target sequences from solution.
Introduction
Biorecognition at a solid/solution interface is a crucial step in many types of sensors. DNA oligonucleotides are frequently used as biorecognition probes, providing selectivity for complementary target nucleic acids that can be used for example to detect and diagnose infectious or genetic diseases, or to study gene expression patterns.(1) The ability of biorecognition probes to bind their complementary target molecule is impacted by surface attachment, which can introduce steric constraints and provide alternative, nonspecific binding opportunities.2,3 Consequently, characterization of surface-bound probe molecules and their ability to bind target molecules from solution has been a major focus in array and sensor literature.
For nucleic acid detection, two factors that help determine the effectiveness of a given system as a sensor are the percentage of surface-bound probes that have captured a target (referred to as the hybridization efficiency) and the number of targets captured (total and per unit area).4−6 Hybridization efficiency is determined under conditions where maximum binding is expected (i.e., high target concentrations, long hybridization times), and provides a means of comparing probe accessibility in different surface configurations, for example, as the probe density or attachment chemistry is changed. A hybridization efficiency of 100% indicates that all of the probes were available for binding to target molecules from solution, while a lower value suggests that some of the probe molecules are unable to perform this function. The number of target molecules captured is important for detection sensitivity, and depends not only on the hybridization efficiency, but also on the probe surface density. Studies investigating hybridization efficiency can lead to improved surfaces for sensing applications, and a number of hybridization efficiency results have been reported for metal surface-immobilized probes.6−16 In general, optimal hybridization efficiencies are achieved for lower probe surface coverages and/or coadsorbed lateral spacer molecules that prevent nonspecific adsorption of probe nucleic acids to the gold substrate.6−11,13,14,17−20 Alternatively, nanostructured surfaces that provide high curvature can facilitate high hybridization efficiency even at high probe surface coverages.9,12,16 Maximizing the number of target molecules bound may or may not coincide with the best conditions for hybridization efficiency; it is not uncommon for more total targets to bind at higher probe densities despite the reduced hybridization efficiency.6,9,10
These determinations all employed linear nucleic acid probes, that is, probes designed specifically not to adopt stable secondary structures. While important for many traditional DNA−DNA binding applications, studies involving strictly linear probes may not be applicable to other systems that employ structured nucleic acid probes, such as molecular beacons, which exhibit a hairpin structure when not bound to target nucleic acid sequences.21−23 Molecular beacon systems are attractive in biosensing for their high specificity, including their ability to discriminate single-base mismatches, and also for in situ analysis without rinsing steps.(21) A number of surface-immobilized molecular beacon detection schemes, both optical24−43 and electrochemical,44−52 have been used in both bioanalytical and biophysical studies of probe−substrate interactions. A number of these works have reported varying probe surface coverage and its impact upon probe surface structure and biosensor response.24,47,49,50 Others have focused on hybridization buffer components and ionic strength or pH effects, finding that these parameters were important to maximize both hairpin unfolding and target binding.24,28,35,38 Differences in probe−surface interactions between structured and unstructured probes have been reported.24,49 These are generally attributable to molecular beacon probe stem-loop secondary structure and include decreased surface density and response to lateral spacer molecules.24,31,32,45,49 Despite these important studies, to our knowledge, there have been no reports quantifying hybridization efficiency for surface-bound hairpin probes.
Hairpin probes immobilized on the surface of metal nanowires offer an opportunity to quantify hybridization efficiency for probes that have been designed to contain secondary structure. When nanowires contain segments of both gold and silver, the striping pattern of these two metals can be used as an identification tag, or barcode, to identify the probe sequence associated with that particle.53,54 We have previously used this approach to perform multiplexed experiments in which several different target sequences were simultaneously detected, each on a different barcoded nanowire.24−27 Here, the nanowire platform facilitates quantification of probe molecules per unit area on the metal surface as well as evaluation of their fluorescence response.24,25 Simple Au nanowires were used as supports for hairpin probes for hybridization efficiency determinations. We note that the nanowire dimensions (ca. 6 μm in length and 300 nm in cross-sectional diameter) are sufficiently large as to be good models for planar surfaces. For <60 nm diameter nanospheres, particle curvature impacts oligonucleotide probe assembly and subsequent target hybridization.55−57 Beyond particle diameters of 100 nm, however, oligonucleotide immobilization behavior has been shown to be essentially the same as that for planar surfaces.(55)
Here, we report the effect of probe surface coverage on the hybridization of hairpin probes bound to Au nanowires. We found that hybridization efficiency was highly dependent on the number of probes present and was maximized at an intermediate coverage. We also investigated the impact of target length on hybridization efficiency and assay performance in order to mimic the response of the sensor to longer, naturally occurring versus synthetic oligonucleotide targets and found that the location of nonhybridizing oligonucleotide stretches influenced performance. Characterization of biosensing platforms employing structured nucleic acids as probes is crucial, as these systems are increasingly used for detection of not only nucleic acids, but multiple other molecular species, such as proteins, small molecules, and ions, via other structured probes such as aptamers.58−60
Results and Discussion
Although the hybridization behavior of surface-bound DNA probes designed to avoid secondary structure has been extensively characterized, to our knowledge, there has been no quantification of hybridization efficiency, much less how it is affected by probe surface coverage, for surface-bound probes exhibiting discrete, intentional secondary structure. We evaluate the effects of (1) probe coverage, (2) oligoethylene glycol diluent spacers, and (3) extra nonhybridizing bases on oligonucleotide target 5′ and 3′ ends on the hybridization efficiency for a hairpin sequence bound to metal nanowires.
The hairpin probe (HP) used here was designed to recognize a 23-nucleotide region of the human immunodeficiency virus (HIV).(25) This region is flanked on either side by five complementary nucleotides to form the stem-loop structure. HP bears a 5′ thiol for attachment to the nanowire and a 3′ TAMRA dye for detection. Emission is quenched due to proximity to the metal surface in the hairpin conformation; when target molecules bind, the TAMRA moves away from the surface and becomes fluorescent.24−27 Experiments were performed on gold scaffolds approximately 6 μm in length and 320 nm in cross-sectional diameter. Thiol attachment of HP to nanowires facilitated control and quantification of the number of HP molecules present per unit area. Fluorescence imaging experiments reported here were mean values from more than 100 individual wires per sample.
Hybridization Efficiency of Surface-Bound Hairpin Probes
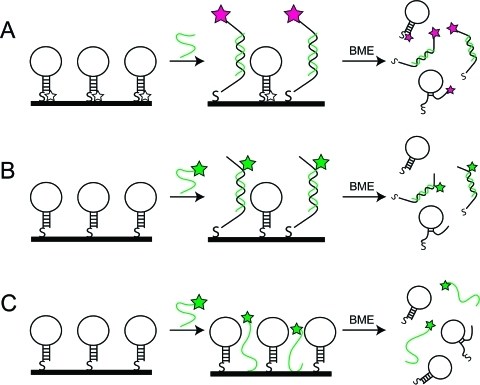
Determination of the hybridization efficiency requires knowledge of the number of molecules of hairpin probe and bound complementary target. Because some target molecules may bind nonspecifically to the surface rather than to their complementary probes, it is also necessary to correct for any nonspecific adsorption. Nonspecific adsorption of target molecules to the metal surface should not contribute to the fluorescence signal, but is undesirable since it would decrease the target molecules available for detection. We measured each of these in separate experiments performed in parallel, as illustrated in Scheme 1. Each experiment quantifies by fluorescence the desired molecule (probe, complementary target, or noncomplementary control) after removal from the nanowire surface by thiol displacement using β-mercaptoethanol (BME). Removal from the nanowires was necessary to avoid complications due to quenching by the gold surface and/or the proximity of the dyes to one another. Additionally, we quantified each component in separate experiments, and omitted the 3′ TAMRA from the HP probe when quantifying the other molecules to avoid complications arising from fluorescence resonance energy transfer.(61) We note that fluorescence strand labeling can impact duplex formation;62,63 however, this is expected to be minimal in our system (e.g., TAMRA has been shown to stabilize DNA duplexes by increasing Tm < 1 °C(63)). The hybridization efficiency was then determined using the following equation: hybridization efficiency = [(labeled target − labeled noncomplementary target)/labeled hairpin] × 100%. Measurements for each type of strand were made in triplicate for determination of standard deviation, and errors were propagated through to obtain uncertainties in hybridization efficiency.
Scheme 1. Quantification of (A) TAMRA-Labeled Hairpin Probe, (B) AlexaFluor488-Labeled Targets, and (C) AlexaFluor488-Labeled Noncomplementary Targets.
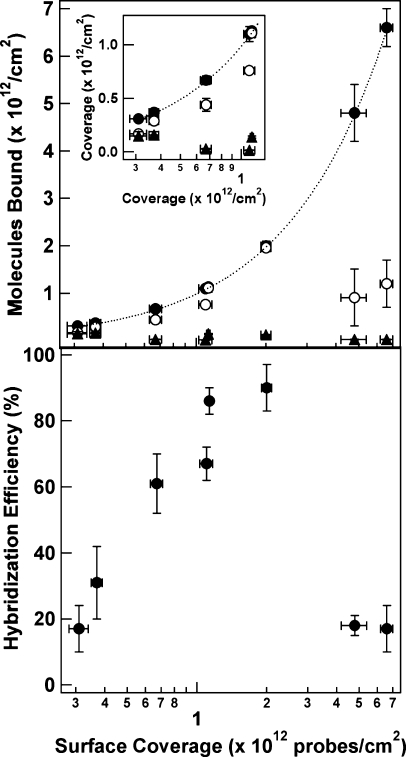
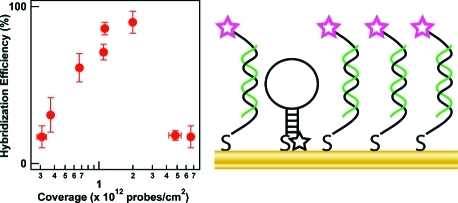
Figure 1 (top) and Supporting Information Table 1 show the effect of hairpin probe surface coverage on the number of complementary or noncomplementary strands bound to the nanowires. HP surface coverage was varied by performing the probe attachment at different ionic strengths to control the screening of the electrostatic repulsions between the adjacent probe molecules.17,24 Higher ionic strengths yielded both higher packing densities and poorer quenching in the absence of target (Supporting Information Figure 1), phenomena that we have observed previously.(24) At ≤1 × 1012 probes/cm2, the number of targets (Tcomp) bound was low, even as compared to the number of probes (Figure 1, top). At intermediate coverage (1.13−2.0 × 1012 probes/cm2), the number of bound target molecules approached the number of probes. Finally, at higher probe surface coverage (4.8−6.6 × 1012 probes/cm2), the number of bound targets per unit area decreased substantially despite the increased number of probe molecules. The total number of noncomplementary target (TNC) molecules bound stayed relatively constant and low (Supporting Information Table 1). The TNC term in the equation for hybridization efficiency was therefore only important at the lowest coverages. Hybridization efficiencies, plotted in Figure 1 (bottom panel) were low at the lowest probe coverages (∼20%), increased to a maximum of ∼90% at intermediate probe coverage, then decreased to ∼20% again at the highest coverages (Figure 1, bottom).
Figure 1.
(top) Effect of HP surface coverage on the number of complementary targets (Tcomp, open circles), and noncomplementary targets (TNC, filled triangles) present. Inset: Expanded view of the lowest coverages. The number of probe strands is also included at each coverage for comparison (HP, filled circles), and the dotted line indicates the target coverage that would correspond to 100% hybridization efficiency, assuming no nonspecific adsorption. (bottom) Effect of HP surface coverage on the hybridization efficiency of HP probes bound to Au nanowires. The bottom axis is shared for both graphs.
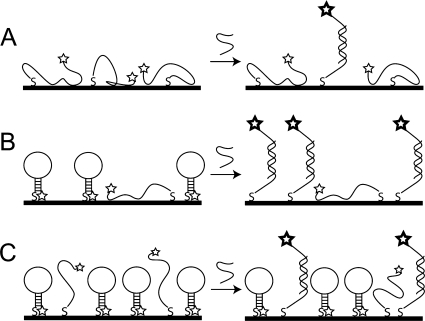
Our interpretation of these data in terms of the DNA−nanowire interface at different coverages is shown in Scheme 2. At low probe coverages, we propose that the probes were unfolded and bound to the metal surface via the nitrogen atoms of the nucleobases.(64) The availability of surface binding sites for nonspecific adsorption and the inaccessibility of the probe sequence for Watson−Crick base pairing with targets contributed to low hybridization efficiency (Scheme 2A). As the probes packed more densely (Scheme 2B), they provided additional steric and electrostatic repulsion to help reduce nonspecific adsorption, and were more accessible to binding target molecules from solution, leading to increased hybridization efficiency. The low hybridization efficiencies seen at the highest probe coverages can be explained by steric hindrance, which also detrimentally affects the hybridization efficiency of unstructured probes at high densities.6−9,11 There were many target binding sites, but targets were either unable to induce unfolding in the hairpins, or could not intercalate into the densely packed probe layer to hybridize to hairpins that might have been unfolded (Scheme 2C).
Scheme 2. Depiction of Hybridization Behavior at (A) Low, (B) Intermediate, and (C) High Hairpin Probe Coverages.
Further insight can be gained by examining the fluorescence intensity for HP-coated nanowires in the presence and absence of target. In the absence of target, the nanowires should appear nonfluorescent, as the fluorophore is in close proximity to the metal surface and emission is quenched.26,65 Upon addition of a complementary target, the nanowires should appear bright, as the hairpin intramolecular 5 bp stem is broken and fluorophore is freed from constraint at the surface. This is largely what we observe, in that the samples to which target has been added are all much brighter than those without target (Supporting Information Figure 1). However, the fluorescence increases with increasing probe coverage even in the absence of target (Supporting Information Figure 2), attributable to a higher overall number of probes. The increase is roughly linear with no substantial changes in slope, indicating that the degree of quenching remains essentially constant as the surface coverage of HP changes. This can be interpreted as either (1) the distribution of HP conformations on the surface is unaffected by HP surface density, or (2) all conformations adopted by HP give essentially the same degree of quenching, such that even if the conformation changes with coverage, the intensity per HP molecule remains constant. We favor the second interpretation, which is physically possible because of the relatively long-range quenching provided by the metal surface, and is consistent with the low nonspecific binding and low hybridization efficiencies we observe at low surface coverage. Quenching can be effective up to 5 nm from the metal surface,26,65,66 which corresponds to 15 base pairs of duplex DNA oriented perpendicular to the surface.
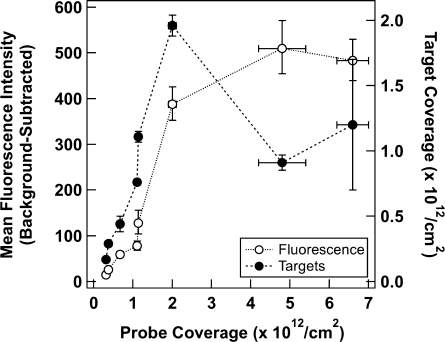
The change in fluorescence intensity upon target binding (i.e., target-present values minus target-absent values) increased with increasing probe coverage up to 5 × 1012/cm2 and then leveled off. This is in contrast to both the hybridization efficiency and the number of target molecules bound per unit area on the nanowire surface, both of which decreased after reaching a maximum at ∼2 × 1012HP/cm2 (Figure 2). We attribute the increased fluorescence intensity despite fewer bound target molecules to restricted gyration of the duplex DNA at high probe packing densities, leading to a greater average distance between the TAMRA and the metal surface and hence greater emission per event than is observed at lower HP coverage. Plaxco and co-workers have observed a similar effect with an electrochemical hairpin system, wherein the signal was highly dependent on both target-bound and target-unbound probe conformations at different densities.(47)
Figure 2.
Background-subtracted fluorescence intensity as determined by microscopy (open circles) and the number of target oligonucleotides present (filled circles) as a function of HP surface coverage. Each fluorescence intensity point corresponds to the mean from >100 individual imaged nanowires. Error bars for fluorescence are 95% confidence intervals; error bars for hybridization efficiency and number of targets present are standard deviations. Lines between points are included to guide the eye.
We note that there is not direct evidence that the molecular beacon probes initially exist in a hairpin structure on the surface; similar changes in fluorescence would be expected just due to changes in the rigidity and hence average surface−dye separation of single- versus double-stranded unstructured probes.(67) It is, however, reasonable to assume that many probes do adopt hairpin structures due to the greater stability of hairpin versus extended structures, and observed differences between traditional unstructured probes and molecular beacon probes support this interpretation.(24) We envision our surface as having a population of some hairpin and some extended conformations coexisting and interconverting, and attempted to show this in Scheme 2. Some intermolecular hybridization between the 3′ and 5′ ends of adjacent strands is also possible.
Effect of Surface Passivation with Thiolated Oligoethylene Glycol on Hybridization Efficiency
A common approach to increasing hybridization efficiency for linear probes is to laterally space them out on the surface by backfilling or coadsorbing the probe with a diluent such as mercaptohexanol or a thiolated oligoethylene glycol.6,8,10,11,17−20,68−73 These molecules are used to prevent multipoint attachment of thiolated probe molecules through their nucleobases, and as such make them more accessible for target binding from solution. They also help protect the surface from nonspecific adsorption of target or other molecules present in the solution. Results for surface dilution of molecular beacon probes using short chain thiols have been less encouraging with regard to quenching efficiency, which was evaluated by fluorescence quantification pre- and post-target addition.24,32 These experiments(24) were performed at relatively high probe coverages of 6.4 × 1012 probes/cm2 pre-spacer addition; if in fact the HPs are unfolded onto the surface at very low coverage, backfilling can be expected to lead to more correct secondary structure and greater availability for target binding, increasing hybridization efficiency. We evaluated the impact of backfilling with a thiolated oligoethylene glycol, HS-OEG, on the hybridization efficiency of nanowire bound HP. We chose this molecule due to the ability of ethylene glycol to reduce biofouling.74,75 Samples were prepared at two assembly ionic strengths (50 and 30 mM buffer) to provide samples of intermediate and low probe surface density. After backfilling with HS-OEG, the samples had 9 ± 1 × 1011 and 3.57 ± 0.08 × 1011 probes/cm2, and hybridization efficiencies of 68 ± 8% and 52 ± 11%, respectively. As compared to samples with similar probe densities that had not been backfilled with HS-OEG, this represented no difference for the higher coverage sample, but a 60% improvement in hybridization efficiency for the lower coverage sample. The improvement in hybridization efficiency for the 3.6−3.7 × 1011 probes/cm2 sample was due to a more than 10-fold decrease in the amount of noncomplementary binding: (1.6 ± 0.3) × 1011TNC/cm2 without HS-OEG spacers and (1.2 ± 0.2) × 1010TNC/cm2 with HS-OEG spacers. Binding of Tcomp was slightly reduced upon spacer inclusion, from (2.9 ± 0.2) × 1011Tcomp/cm2 to (2.0 ± 0.4) × 1011Tcomp/cm2. These data are consistent with our hypothesis that HPs interacted with the metal surface along their lengths at the lowest surface coverages (Scheme 2, top); the HS-OEG molecules prevented this interaction and resulted in a greater percentage of HPs adopting the desired secondary structure. Our data cannot differentiate Tcomp strands associated with the probes from those bound nonspecifically to the sites on the metal surface where TNC bound in the absence of HS-OEG; presumably both occurred. Furthermore, it is likely that the amount of nonspecific adsorption to the metal was very similar for Tcomp and TNC, which are similar in length and percent purine/pyrimidine composition. If the HP probes were fully available for hybridization at the low coverages without HS-OEG, we would expect the Tcomp signal to be the sum of hybridized and nonspecifically adsorbed strands, and the TNC signal to indicate the amount of nonspecific adsorption to the metal surface. Thus, Tcomp should have decreased substantially when it was restricted to binding only through hybridization to HP and not to the bare Au between the probes. Tcomp decreased only slightly, suggesting that the reduction in nonspecific adsorption was offset by an increase in hybridization.
Hybridization Efficiency of Longer Targets
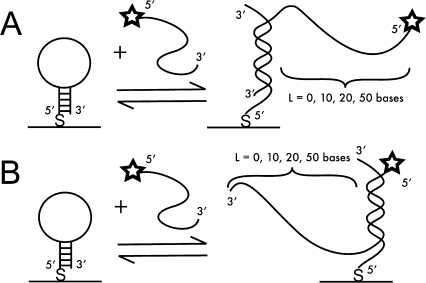
In a real-world biosensing application, target length will not be matched to the length probe’s hairpin loop, but will most likely be much longer, i.e., hundreds of bases. The placement of a complementary target sequence within a longer strand of nucleic acid can be expected to impact its hybridization to a surface-bound probe due to steric hindrance.9,76 We therefore evaluated the effect of targets having additional, nonhybridizing nucleotides on either the 5′ or 3′ end to gain insight into the hybridization behavior at the solution−surface interface using a controlled system. It is not always possible to control the location of the complementary portion of the target sequence in real samples; however, when polymerase chain reaction (PCR) amplification is performed prior to detection, the sequence at the 3′ or 5′ end of the amplicon can be used.
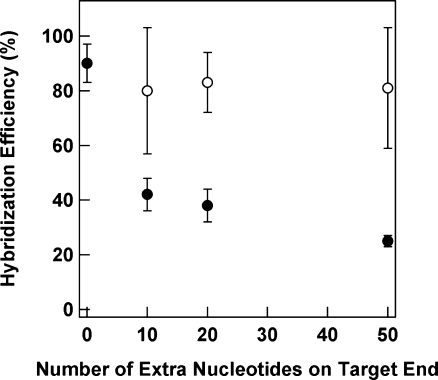
Targets of extended length were employed that represent the region complementary to the hairpin loop, plus an extra 10, 20, and 50 nucleotides on either the 5′ or 3′ end of the HIV sequence (Scheme 3). Noncomplementary targets used in these assays represented matched lengths taken from an Influenza B sequence. The effects of these extra bases on hybridization efficiency are presented in Table 1 and Figure 3. Note that although HP attachment was performed under the same conditions for each target, because the nanowires used for the 5′ target experiments were slightly longer than those used for the 3′ targets, HP coverage differed between the two (Table 1). Thus, the change in hybridization efficiency for the 10 nt overhang as compared to the fully matched target cannot be attributed to the additional length alone, and may be primarily due to differences in probe coverage. However, we are able to directly compare the effect of adding 10, 20, versus 50 nt on either the 3′ or 5′ end. There was little change in hybridization efficiency with increasing length for targets incorporating extra 5′ nt, with values remaining near 80% even for the longest target tested. The 3′ overhang had a greater effect, decreasing from ∼40% for 10 nt to just 25% for 50 nt, which is consistent with the greater steric hindrance associated with the increased length on the 3′ end, which is oriented toward the nanowire surface (Scheme 3). These results are consistent with those of Fan and co-workers, who also observed a higher extent of hybridization when extra nucleotides of PCR amplicons were oriented toward solution instead of gold electrode surfaces.(73)
Scheme 3. Illustration of Hybridization of Fluorescently Labeled Targets Incorporating Additional Nonhybridizing Nucleotides on the 5′ End (A), or 3′ End (B) of the Region Complementary to Surface-Bound Hairpin Probes.
Table 1. Surface Coverages, Footprints, and Hybridization Efficiencies for Hybridization Assays Incorporating Targets with Additional Nonhybridizing Nucleotides.
| extra nucleotide location and length | HP surface coverage (× 1012 probes/cm2) | complementary target coverage (× 1012/cm2) | noncomplementary target coverage (× 1012/cm2) | hybridization efficiency (%) |
|---|---|---|---|---|
| nonea | 2.0 ± 0.1 | 1.96 ± 0.08 | 0.12 ± 0.01 | 90 ± 7 |
| nonea | 4.8 ± 0.6 | 0.91 ± 0.06 | 0.021 ± 0.009 | 18 ± 3 |
| 5′-10 nt | 2.8 ± 0.6 | 2.3 ± 0.4 | 0.02 ± 0.01 | 80 ± 23 |
| 5′-20 nt | 2.4 ± 0.2 | 2.0 ± 0.2 | 0.025 ± 0.005 | 83 ± 11 |
| 5′-50 nt | 2.6 ± 0.5 | 2.1 ± 0.4 | 0.03 ± 0.02 | 81 ± 22 |
| 3′-10 nt | 3.5 ± 0.3 | 1.5 ± 0.2 | 0.015 ± 0.005 | 42 ± 6 |
| 3′-20 nt | 3.6 ± 0.4 | 1.4 ± 0.2 | 0.007 ± 0.003 | 38 ± 6 |
| 3′-50 nt | 3.6 ± 0.3 | 0.89 ± 0.05 | 0.0159 ± 0.0009 | 25 ± 2 |
Because hybridization efficiency is dependent on probe coverage, two values for the fully matched target Tcomp are included to bracket the HP coverage values found in the experiments using longer targets.
Figure 3.
Effect of target length on the hybridization efficiency of HPs bound to metal nanowires. Targets had extra nonhybridizing nucleotides on the 5′ end (open circles) or 3′ end (closed circles) of the region complementary to the probe sequence and were labeled at the 5′ end with AlexaFluor 488 to enable quantification. Error bars represent the standard deviation in the hybridization efficiency measurement.
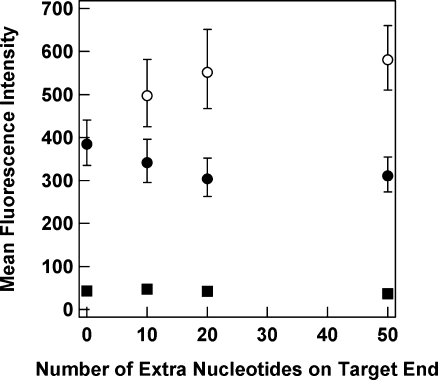
Fluorescence intensity data for experiments in which TAMRA-labeled HP-coated nanowires were exposed to the longer unlabeled complementary and noncomplementary targets are shown in Figure 4. These experiments were performed using a single set of HP-coated nanowires ((2−3) × 1012HP/cm2) and hence can be directly compared. Complementary targets of all lengths gave substantially higher fluorescence intensity than noncomplementary controls. For targets incorporating extra nucleotides on the 5′ end, fluorescence increased somewhat as compared to unmodified Tcomp (p < 0.005 for the increase from 0 to 10 nt on the 5′ end). This may be due to the bound targets interacting with neighboring probes. The fluorescence intensity was however not strongly dependent on length; values for 10, 20, or 50 additional nucleotides were not statistically different (p > 0.1). Additional nt on the 3′ end resulted in a slight decrease in fluorescence intensity with increasing length (p < 0.05 for 0 vs 20 nt on 3′ end), consistent with the lower number of bound targets (Table 1). However, this decrease was also not statistically significant when comparing 10, 20, and 50 additional nt targets to each other (p > 0.1). These data were acquired in buffer solutions under high target concentrations (1 μM) and are hence not directly relevant to bioassay performance. They are encouraging for surface-immobilized hairpin bioassays because they indicate that extra nonhybridizing nucleotides did not appreciably hinder target binding, and the fluorescence response retained specificity even when challenged with long noncomplementary targets.
Figure 4.
The effect of target length on fluorescence hybridization signal for TAMRA-labeled HPs bound to metal nanowires. Targets were unlabeled and had extra nonhybridizing nucleotides on the 5′ end (open circles) or 3′ end (closed circles) of the region complementary to the probe sequence. Noncomplementary sequences of different lengths are shown for comparison (closed squares). Each fluorescence intensity point corresponds to mean from >100 individual imaged nanowires. Error bars represent the 95% confidence level.
Conclusions
We report the first determination of hybridization efficiency for hairpin nucleic acid probes immobilized on a surface. The hybridization efficiency was highly dependent on probe coverage and reached a maximum of ∼90% at a coverage of 1.13−2.0 × 1012 probes/cm2. Hybridization efficiencies dropped to ∼20% at both higher and lower coverages, due to steric hindrance and adsorption of the probe to the metal surface, respectively. The use of thiolated oligoethylene glycol spacers increased the hybridization efficiency at the lowest coverages by displacing nonspecifically bound nucleotides from the surface. Inclusion of extra nucleotides at the 5′ end of the target resulted in retention of high hybridization efficiency and fluorescence intensity with respect to target length, whereas extra nucleotides on the target 3′ end yielded a decrease in hybridization efficiency, but only a slight reduction in fluorescence intensity with respect to target length. These investigations into the behavior of surface-bound structured nucleic acid probes can serve as guidelines in the interfacial design of emerging technologies that employ structured probes, such as those based on molecular beacon or aptamer probes on nanospheres, nanowires, and/or planar supports.
Materials and Methods
Materials
All chemicals and biochemicals were used as received without further purification except where noted. Water for all experiments was purified using a Barnstead Nanopure system to a resistivity of 18 MΩ. Buffers employed in this work were (1) 1 M PBS (1 M NaCl; 10 mM sodium phosphate, pH 7.2), (2) 0.3 M PBS (0.3 M NaCl; 10 mM sodium phosphate, pH 7.2), (3) 10 mM PBS (10 mM sodium phosphate, pH 7.2), and (4) 0.5 M CAC (0.5 M NaCl; 20 mM cacodylic acid; 0.5 mM EDTA, pH 7.0).25,31
Au nanowires were fabricated as previously reported and suspended to a concentration of 1 × 109 wires/mL in ethanol.53,77,78 Wire dimensions were 5.4−5.7 μm in length and 320 nm in cross-sectional diameter.53,77,78 The hairpin DNA probe sequence was designed as described elsewhere and modified with a 5′ thiol for conjugation to Au nanowires and a 3′ TAMRA dye for fluorescence elucidation.(24) All DNA sequences (Table 2) were obtained from Integrated DNA Technologies, Inc. (Coralville, IA). Thiolated hairpin probes were received as disulfides and were cleaved before use to yield terminal 5′ thiols. Cleavage was performed via reaction of 100 μM DNA with 100 mM dithiothreitol (DTT) in 0.1 M phosphate buffer, pH 8.3, for 30 min. The resultant product was then purified by a Centrispin 10 column (Princeton Separations) according to manufacturer protocols.
Table 2. Sequences Used in This Work.
| sequence (5′ to 3′) | comments |
|---|---|
| thiol(CH2)6-GCG AGT GTT AAA AGA GAC CAT CAA TGA GCT CGC-TAMRAa | HP; Hairpin probe specific for a 23-base region of HIVb |
| CTC ATT GAT GGT CTC TTT TAA CAc | Tcomp; Target strand complementary to loop region of HP |
| Alexa488-TCA CCG GTT CCG CAG ACC ACT ATG | TNC; Noncomplementary control target strandb,d |
| TGG AAT AGG CCC TGC ATG TAC TGG ATG TAA CCT GTC CCA TTC TGC AGC TTC CTC ATT GAT GGT CTC TTT TAA CAT TTG CAT TGC TGC CTG ATG TCC CCC CAC TGT GTT TAG CAT AGT ATT TAA Gc | Extended target sequence showing nonhybridizing nucleotides on either side of (underlined) Tcomp region of the HIV sequence; our experiments used from 10 to 50 nt on 3′ or 5′ end, as noted in the text and figuresb |
| CAA TAG GTC TTT CCA CTG CAA ACA CTG GGC TGC AGC TTA TTT GGC CAG ACC CTC CGT CTC CAC CTA CTT CGT Tc | Noncomplementary target control representing a 73-base region of Influenza B virus; here, the underlined region is complementary to a probe used in reference (79) and was used as the central segment onto which 5′ nt were added to produce noncomplementary targets of desired length |
The italicized portions of probe sequence indicate regions of self-complementarity.
HIV, HCV, and Influenza B are all RNA viruses; however, here, our synthetic DNA targets served as viral nucleic acid mimics.
These sequences were available either unmodified or modified with a 5′ AlexaFluor 488 for hybridization efficiency determination.
Representative of a 23-nucleotide sequence of Hepatitis C Virus.
Determination of Surface Coverage
Oligonucleotide sequences were displaced from the metal nanowire surfaces by treatment with an excess of BME.12,24 Briefly, 2.5 × 107 DNA-coated nanowires were suspended in 195 μL of 0.5 M CAC, and 5 μL of BME was introduced into the tubes for overnight reaction in the dark. Nanowires were pelleted by centrifugation, and the supernatant fluorescence was quantified using a Fluorolog-3 fluorimeter (TAMRA parameters: λex = 558 nm, λem = 580 nm; AlexaFluor488 parameters: λex = 492 nm, λem = 517 nm). Calibration curves were generated for each type of fluorescent DNA strand, and surface coverages were calculated based on nanowire dimensions.
Determination of Hybridization Efficiency
Twenty-five microliters of nanowires (2.5 × 107 wires) were rinsed one time by centrifugation (1 min at 7700g) out of ethanol, two times in 25 μL of water, and one time in 50 μL of selected buffer before resuspension in 75 μL of buffer to be used in the attachment process. To vary the surface coverage, buffers with differing ionic strengths were used. A high ionic strength buffer, 1 M PBS, was diluted with 10 mM PBS. To obtain buffers of very low ionic strengths, solutions of 10 mM PBS were diluted with deionized (DI) water. HP DNA was added such that the final concentration was equal to 1.0 μM. Samples were prepared in triplicate in order to generate standard deviations for each measurement. After reaction for 30 min, each sample was rinsed by centifugation two times with 75 μL of buffer. The nanowires bearing the HP probes were hybridized with fluorescently labeled targets from Table 2 as noted in the text, at target concentrations of 1.0 μM and volumes of 333 μL in 0.5 M CAC hybridization buffer. Posthybridization, each sample was washed two times with 333 μL of 0.5 M CAC, and the surface coverage of each type of labeled strand was quantified as described above.
After attachment of HPs to Au wires as detailed above in either 50 mM PBS or 30 mM PBS, the DNA-coated wires were exposed to 1 mM HS(CH2)11(OCH2CH2)3OH, (referred to as HS-OEG, SensoPath Technologies, Bozeman, MT) in 75 μL of either 50 mM PBS or 30 mM PBS for 30 min. Samples were then rinsed two times with 75 μL of 50 mM PBS or 30 mM PBS and hybridized as previously described, using labeled/unlabeled Tcomp and TNC as the target and fluorescently labeled noncomplementary target, respectively.
Optical Microscopy
For samples to be imaged under an optical microscope, HP probes were immobilized to nanowires as described above, and these nanowires were then hybridized with unlabeled targets from Table 2 as noted in the text, at target concentrations of 1.0 μM. Targets were not rinsed out posthybridization.(25) Directly after hybridization, a 10 μL aliquot of nanowire suspension was placed onto a glass coverslip. Images were acquired using a Nikon TE-300 inverted microscope with a CFI plan fluor 60x oil immersion lens (N.A. = 1.4) and ImagePro Plus version 4.5 and 7.0 software. Images were captured using a Coolsnap HQ camera (Photometrics). The light source was a 300 W ozone-free Xe lamp. A Nikon filter cube with Semrock filters specific for TAMRA excitation and emission (excitation: 531/40, emission: 593/40, dichroic: 562 long-pass) was employed to obtain fluorescence images, and reflectance images were obtained with white light. NBSee Software (Nanoplex Technologies) was employed to quantify fluorescence intensity.(80) Briefly, the reflectance and corresponding fluorescence images were analyzed for several hundred nanowires, and a log mean fluorescence intensity was generated.(25)
Acknowledgments
This work was funded by the National Institutes of Health (R01 EB000268 and R21 CA137595).
Supporting Information Available
Raw numerical data for the populations of hairpins, complementary targets, and noncomplementary targets to determine hybridization efficiency, quantification of fluorescence intensity in the presence and absence of target strands at different probe coverages, fluorescence in the absence of target as a function of probe coverage. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Cantor C. R.; Smith C. L.. Genomics: The Science and Technology Behind the Human Genome Project; Wiley and Sons: New York, 1999. [Google Scholar]
- DNA Microarrays: A Practical Approach; Schena M., Ed.; Oxford University Press: New York, 1999. [Google Scholar]
- Sassolas A.; Leca-Bouvier B. D.; Blum L. DNA Biosensors and Microarrays. Chem. Rev. 2008, 108, 109–139. [DOI] [PubMed] [Google Scholar]
- Ekins R. P.; Chu F. Developing Multianalyte Assays. Trends Biotechnol. 1994, 12, 89–94. [DOI] [PubMed] [Google Scholar]
- Dandy D. S.; Wu P.; Grainger D. W. Array Feature Size Influences Nucleic Acid Surface Capture in DNA Microarrays. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 8223–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson A. W.; Heaton R. J.; Georgiadis R. M. The Effect of Surface Probe Density on DNA Hybridization. Nucleic Acids Res. 2001, 29, 5163–5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelino K.; Kanna B.; Majumdar A. Characterization of Grafting Density and Binding Efficiency of DNA and Proteins on Gold Surfaces. Langmuir 2005, 21, 1956–1961. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y.; Nguyen P.-C. T.; Grainger D. W.; Gamble L. J.; Castner D. G. Structure and DNA Hybridization Properties of Mixed Nucleic Acid/Maleimide−Ethylene Glycol Monolayers. Anal. Chem. 2007, 79, 4390–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicewarner-Peña S. R.; Raina S.; Goodrich G. P.; Fedoroff N. V.; Keating C. D. Hybridization and Extension of Au Nanoparticle-Bound Oligonucleotides. J. Am. Chem. Soc. 2002, 124, 7314–7323. [DOI] [PubMed] [Google Scholar]
- Gong P.; Lee C.-Y.; Gamble L. J.; Castner D. G.; Grainger D. W. Hybridization Behavior of Mixed DNA-Alkylthiol Monolayers on Gold: Characterization by Surface Plasmon Resonance and 32P Radiometric Assay. Anal. Chem. 2006, 78, 3326–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel A. B.; Herne T. M.; Tarlov M. J. Electrochemical Quantitation of DNA Immobilized on Gold. Anal. Chem. 1998, 70, 4670–4677. [DOI] [PubMed] [Google Scholar]
- Demers L. M.; Mirkin C. A.; Mucic R. C.; Reynolds R. A. III; Letsinger R. L.; Elghanian R.; Viswanadham V. A Fluorescence-Based Method for Determining the Surface Coverage and Hybridization Efficiency of Thiol-Capped Oligonucleotides Bound to Gold Thin Films and Nanoparticles. Anal. Chem. 2000, 72, 5535–5541. [DOI] [PubMed] [Google Scholar]
- Boncheva M.; Scheibler L.; Lincoln P.; Vogel H.; Åkerman B. Design of Oligonucleotide Arrays at Interfaces. Langmuir 1999, 15, 4317–4320. [Google Scholar]
- Zu Y.; Gao Z. Facile and Controllable Loading of Single-Stranded DNA on Gold Nanoparticles. Anal. Chem. 2009, 81, 8523–8528. [DOI] [PubMed] [Google Scholar]
- Mirmomtaz E.; Castronovo M.; Grunwald C.; Bano F.; Scaini D.; Ensafi A. A.; Scoles G.; Casalis L. Quantitative Study of the Effect of Coverage on the Hybridization Efficiency of Surface-Bound DNA Nanostructures. Nano Lett. 2008, 8, 4134–4139. [DOI] [PubMed] [Google Scholar]
- Bin X.; Sargent E. H.; Kelley S. O. Nanostructuring of Sensors Determines the Efficiency of Biomolecular Capture. Anal. Chem. 2010, 82, 5928–5931. [DOI] [PubMed] [Google Scholar]
- Herne T. M.; Tarlov M. J. Characterization of DNA Probes Immobilized on Gold Surfaces. J. Am. Chem. Soc. 1997, 119, 8916–8920. [Google Scholar]
- Levicky R.; Herne T. M.; Tarlov M. J.; Satija S. K. Using Self-Assembly to Control the Structure of DNA Monolayers on Gold: A Neutron Reflectivity Study. J. Am. Chem. Soc. 1998, 120, 9787–9792. [Google Scholar]
- Cho Y.-K.; Kim S.; Kim Y. A.; Lim H. K.; Lee K.; Yoon D.; Lim G.; Pak Y. E.; Ha T. H.; Kim K. Characterization of DNA Immobilization and Subsequent Hybridization Using In Situ Quartz Crystal Microbalance, Fluorescence Spectroscopy, and Surface Plasmon Resonance. J. Colloid Interface Sci. 2004, 278, 44–52. [DOI] [PubMed] [Google Scholar]
- Peterson A. W.; Wolf L. K.; Georgiadis R. M. Hybridization of Mismatched or Partially Matched DNA at Surfaces. J. Am. Chem. Soc. 2002, 124, 14601–14607. [DOI] [PubMed] [Google Scholar]
- Tyagi S.; Kramer F. R. Molecular Beacons: Probes that Fluoresce Upon Hybridization. Nat. Biotechnol. 1996, 14, 303–308. [DOI] [PubMed] [Google Scholar]
- Tan W.; Wang K.; Drake T. J. Molecular Beacons. Curr. Opin. Chem Biol. 2004, 8, 547–553. [DOI] [PubMed] [Google Scholar]
- Broude N. E. Stem-Loop Oligonucleotides: A Robust Tool for Molecular Biology and Biotechnology. Trends Biotechnol. 2002, 6, 249–256. [DOI] [PubMed] [Google Scholar]
- Cederquist K. B.; Golightly R. S.; Keating C. D. Molecular Beacon−Metal Nanowire Interface: Effect of Probe Sequence and Surface Coverage on Sensor Performance. Langmuir 2008, 24, 9162–9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoermer R. L.; Cederquist K. B.; McFarland S. K.; Sha M. Y.; Penn S. G.; Keating C. D. Coupling Molecular Beacons to Barcoded Metal Nanowires for Multiplexed, Sealed Chamber DNA Bioassays. J. Am. Chem. Soc. 2006, 128, 16892–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoermer R. L.; Keating C. D. Distance-Dependent Emission from Dye-Labeled Oligonucleotides and Striped Au/Ag Nanowires: Effect of Secondary Structure and Hybridization Efficiency. J. Am. Chem. Soc. 2006, 128, 13243–13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha M. Y.; Yamanaka M.; Walton I. D.; Norton S. M.; Stoermer R. L.; Keating C. D.; Natan M. J.; Penn S. G. Encoded Metal Nanoparticle-Based Molecular Beacons for Multiplexed Detection of DNA. NanoBiotechnology 2005, 1, 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.; Li J.; Liu H.; Liu Q.; Mei Q.; Wang Y.; Zhu J.; He N.; Lu Z. Label-Free Hybridization Detection of a Single Nucleotide Mismatch by Immobilization of Molecular Beacons on an Agarose Film. Nucleic Acids Res. 2002, 30, e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubertret B.; Calame M.; Libchaber A. J. Single-Mismatch Detection Using Gold-Quenched Fluorescent Oligonucleotides. Nat. Biotechnol. 2001, 19, 365–370. [DOI] [PubMed] [Google Scholar]
- Du H.; Disney M. D.; Miller B. L.; Krauss T. D. Hybridization-Based Unquenching of DNA Hairpins on Au Surfaces: Prototypical “Molecular Beacon” Biosensors. J. Am. Chem. Soc. 2003, 125, 4012–4013. [DOI] [PubMed] [Google Scholar]
- Du H.; Strohsahl C. M.; Camera J.; Miller B. L.; Krauss T. D. Sensitivity and Specificity of Metal Surface-Immobilized “Molecular Beacon” Biosensors. J. Am. Chem. Soc. 2005, 127, 7932–7940. [DOI] [PubMed] [Google Scholar]
- Peng H.-I.; Strohsahl C. M.; Leach K. E.; Krauss T. D.; Miller B. L. Label-free DNA Detection on Nanostructured Ag Surfaces. ACS Nano 2009, 3, 2265–2273. [DOI] [PubMed] [Google Scholar]
- Brown L. J.; Cummins J.; Hamilton A.; Brown T. Molecular Beacons Attached to Glass Beads Fluoresce Upon Hybridisation to Target DNA. Chem. Commun. 2000, 621–622. [Google Scholar]
- Situma C.; Moehring A. J.; Noor M. A. F.; Soper S. A. Immobilized Molecular Beacons: A New Strategy Using UV-Activated Poly(methyl methacrylate) Surfaces to Provide Large Fluorescence Sensitivities for Reporting on Molecular Association Events. Anal. Biochem. 2007, 363, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G.; Tan W. Molecular-Beacon-Based Array for Sensitive DNA Analysis. Anal. Biochem. 2004, 331, 216–223. [DOI] [PubMed] [Google Scholar]
- Yao G.; Hang X.; Yokota H.; Yanagida T.; Tan W. Monitoring Molecular Beacon DNA Probe Hybridization at the Single-Molecule Level. Chem.—Eur. J. 2003, 9, 5686–5692. [DOI] [PubMed] [Google Scholar]
- Liu X.; Farmerie W.; Schuster S.; Tan W. Molecular Beacons for DNA Biosensors with Micrometer to Submicrometer Dimensions. Anal. Biochem. 2000, 283, 56–63. [DOI] [PubMed] [Google Scholar]
- Liu X.; Tan W. A Fiber-Optic Evanescent Wave DNA Biosensor Based on Novel Molecular Beacons. Anal. Chem. 1999, 71, 5054–5059. [DOI] [PubMed] [Google Scholar]
- Fang X.; Liu X.; Schuster S.; Tan W. Designing a Novel Molecular Beacon for Surface-Immobilized DNA Hybridization Studies. J. Am. Chem. Soc. 1999, 121, 2921–2922. [Google Scholar]
- Yang R.; Jin J.; Chen Y.; Shao N.; Kang H.; Xiao Z.; Tang Z.; Wu Y.; Zhu Z.; Tan W. Carbon Nanotube-Quenched Fluorescent Oligonucleotides: Probes that Fluoresce Upon Hybridization. J. Am. Chem. Soc. 2008, 130, 8351–8358. [DOI] [PubMed] [Google Scholar]
- Mao X.; Xu M.; Zeng Q.; Zeng L.; Liu G. Molecular Beacon-Functionalized Gold Nanoparticles as Probes in Dry-Reagent Strip Biosensor for DNA Analysis. Chem. Commun. 2009, 3065–3067. [DOI] [PubMed] [Google Scholar]
- Song S.; Liang Z.; Zhang J.; Wang L.; Li G.; Fan C. Gold-Nanoparticle-Based Multicolor Nanobeacons for Sequence-Specific DNA Analysis. Angew. Chem., Int. Ed. 2009, 48, 8670–8674. [DOI] [PubMed] [Google Scholar]
- Jayagopal A.; Halfpenny K. C.; Perez J. W.; Wright D. W. Hairpin DNA-Functionalized Gold Colloids for the Imaging of mRNA in Live Cells. J. Am. Chem. Soc. 2010, 132, 9789–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C.; Plaxco K. W.; Heeger A. J. Electrochemical Interrogation of Conformational Changes as a Reagentless Method for the Sequence-Specific Detection of DNA. Proc. Natl. Acad. Sci. U.S.A. 2003, 100, 9134–9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin A. A.; Lai R. Y.; Baker B. R.; Heeger A. J.; Plaxco K. W. Sequence-Specific, Electronic Detection of Oligonucleotides in Blood, Soil, and Foodstuffs with the Reagentless, Reusable E-DNA Sensor. Anal. Chem. 2006, 78, 5671–5677. [DOI] [PubMed] [Google Scholar]
- Lai R. Y.; Lagally E. T.; Lee S.-H.; Soh H. T.; Plaxco K. W.; Heeger A. J. Rapid, Sequence-Specific Detection of Unpurified PCR Amplicons via a Reusable, Electrochemical Sensor. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 4017–4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci F.; Lai R. Y.; Heeger A. J.; Plaxco K. W.; Sumner J. J. Effect of Molecular Crowding on the Response of an Electrochemical DNA Sensor. Langmuir 2007, 23, 6827–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin A. A.; Hunt B. V. S.; White R. J.; Plaxco K. W. Effects of Probe Length, Probe Geometry, and Redox-Tag Placement on the Performance of the Electrochemical E-DNA Sensor. Anal. Chem. 2009, 81, 2150–2158. [DOI] [PubMed] [Google Scholar]
- Steichen M.; Buess-Herman C. Electrochemical Detection of the Immobilization and Hybridization of Unlabeled Linear and Hairpin DNA on Gold. Electrochem. Commun. 2005, 7, 416–420. [Google Scholar]
- Wang X.; Yun W.; Dong P.; Zhou J.; He P.; Fang Y. A Controllable Solid-State Ru(bpy)32+ Electrochemiluminescence Film Based on Conformation Change of Ferrocene-Labeled DNA Molecular Beacon. Langmuir 2008, 24, 2200–2205. [DOI] [PubMed] [Google Scholar]
- Kjällman T. H. M.; Peng H.; Soeller C.; Travas-Sejdic J. Effect of Probe Density and Hybridization Temperature on the Response of an Electrochemical Hairpin-DNA Sensor. Anal. Chem. 2008, 80, 9460–9466. [DOI] [PubMed] [Google Scholar]
- Gong H.; Zhong T.; Gao L.; Li X.; Bi L.; Kraatz H.-B. Unlabeled Hairpin DNA Probe for Electrochemical Detection of Single-Nucleotide Mismatches Based on MutS−DNA Interactions. Anal. Chem. 2009, 81, 8639–8643. [DOI] [PubMed] [Google Scholar]
- Nicewarner-Peña S. R.; Freeman G. P.; Reiss B. D.; He L.; Peña D. J.; Walton I. D.; Cromer R.; Keating C. D.; Natan M. J. Submicrometer Metallic Barcodes. Science 2001, 294, 137–141. [DOI] [PubMed] [Google Scholar]
- Brunker S. E.; Cederquist K. B.; Keating C. D. Metallic Barcodes for Multiplexed Bioassays. Nanomedicine 2007, 2, 695–710. [DOI] [PubMed] [Google Scholar]
- Hill H. D.; Millstone J. E.; Banholzer M. J.; Mirkin C. A. The Role Radius of Curvature Plays in Thiolated Oligonucleotide Loading on Gold Nanoparticles. ACS Nano 2009, 3, 418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist K. B.; Keating C. D. Curvature Effects in DNA:Au Nanoparticle Conjugates. ACS Nano 2009, 3, 256–260. [DOI] [PubMed] [Google Scholar]
- Kira A.; Kim H.; Yasuda K. Contribution of Nanoscale Curvature to Number Density of Immobilized DNA on Gold Nanoparticles. Langmuir 2009, 25, 1285–1288. [DOI] [PubMed] [Google Scholar]
- Famulok M.; Hartig J. S.; Mayer G. Functional Aptamers and Aptazymes in Biotechnology, Diagnostics, and Therapy. Chem. Rev. 2007, 107, 3715–3743. [DOI] [PubMed] [Google Scholar]
- Functional Nucleic Acids for Analytical Applications; Li Y., Lu Y., Eds.; Integrated Analytical Systems; Springer Publishing: New York, 2009. [Google Scholar]
- The Aptamer Handbook; Klussmann S., Ed.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006. [Google Scholar]
- Lakowicz J. R.Principles of Fluorescence Spectroscopy, 3rd ed.; Springer Publishing: New York, 2006; pp 443−475. [Google Scholar]
- Zhang L.; Hurek T.; Reinhold-Hurek B. Position of the Fluorescent Label Is A Crucial Factor Determining Signal Intensity in Microarray Hybridizations. Nucleic Acids Res. 2005, 33, e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira B. G.; You Y.; Behlke M. A.; Owczarzy R. Effect of Fluorescent Dyes, Quenchers, and Dangling Ends on DNA Duplex Stability. Biochem. Biophys. Res. Commun. 2005, 327, 473–484. [DOI] [PubMed] [Google Scholar]
- Opdahl A.; Petrovych D. Y.; Kimura-Suda H.; Tarlov M. J.; Whitman L. J. Independent Control of Grafting Density and Conformation of Single-Stranded DNA Brushes. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersten J.; Nitzan A. Spectroscopic Properties of Molecules Interacting with Small Dielectric Particles. J. Chem. Phys. 1981, 75, 1139–1152. [Google Scholar]
- Lakowicz J. R. Radiative Decay Engineering: Biophysical and Biomedical Applications. Anal. Biochem. 2001, 298, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell D. J.; Taylor J. R.; Nie S. Self-Assembled Nanoparticle Probes for Recognition and Detection of Biomolecules. J. Am. Chem. Soc. 2002, 124, 9606–9612. [DOI] [PubMed] [Google Scholar]
- Peterlinz K. A.; Georgiadis R. M. Observation of Hybridization and Dehybridization of Thiol-Tethered DNA Using Two-Color Surface Plasmon Resonance Spectroscopy. J. Am. Chem. Soc. 1997, 119, 3401–3402. [Google Scholar]
- Peeters S.; Stakenborg T.; Reekmans G.; Laureyn W.; Lagae L.; Van Aerschot A.; Van Ranst M. Impact of Spacers on the Hybridization Efficiency of Mixed Self-Assembled DNA/Alkanethiol Films. Biosens. Bioelectron. 2008, 24, 72–77. [DOI] [PubMed] [Google Scholar]
- Steichen M.; Brouette N.; Buess-Herman C.; Fragneto G.; Sferrazza M. Interfacial Behavior of a Hairpin DNA Probe Immobilized on Gold Surfaces. Langmuir 2009, 25, 4162–4167. [DOI] [PubMed] [Google Scholar]
- Peled D.; Daube S. S.; Naaman R. Selective Enzymatic Labeling to Detect Packing-Induced Denaturation of Double-Stranded DNA at Interfaces. Langmuir 2008, 24, 11842–11846. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y.; Gamble L. J.; Grainger D. W.; Castner D. G. Mixed DNA/Oligo(ethylene glycol) Functionalized Gold Surfaces Improve DNA Hybridization in Complex Media. Biointerphases 2006, 1, 82–92. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Lao R.; Song S.; Yan Z.; Fan C. Design of an Oligonucleotide-Incorporated Nonfouling Surface and Its Application in Electrochemical DNA Sensors for Highly Sensitive and Sequence-Specific Detection of Target DNA. Anal. Chem. 2008, 80, 9029–9033. [DOI] [PubMed] [Google Scholar]
- Zareie H. M.; Boyer C.; Bulmus V.; Nateghi E.; Davis T. P. Temperature-Reponsive Self-Assembled Monolayers of Oligo(ethylene glycol): Control of Biomolecular Recognition. ACS Nano 2008, 2, 757–765. [DOI] [PubMed] [Google Scholar]
- Feldman K.; Hähner G.; Spencer N. D.; Harder P.; Grunze M. Probing Resistance to Protein Adsorption of Oligo(ethylene glycol)-Terminated Self-Assembled Monolayers by Scanning Force Microscopy. J. Am. Chem. Soc. 1999, 121, 10134–10141. [Google Scholar]
- Stedtfeld R. D.; Wick L. M.; Baushke S. W.; Tourlousse D. M.; Herzog A. B.; Xia Y.; Rouillard J. M.; Klappenbach J. A.; Cole J. R.; Gulari E.; Tiedje J. M.; Hashsham S. A. Influence of Dangling Ends and Surface-Proximal Tails of Targets on Probe−Target Duplex Formation in 16S rRNA Gene-Based Diagnostic Arrays. Appl. Environ. Microbiol. 2007, 73, 380–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating C. D.; Natan M. J. Striped Metal Nanowires as Building Blocks and Optical Tags. Adv. Mater. 2003, 15, 451–454. [Google Scholar]
- Reiss B. D.; Freeman R. G.; Walton I. D.; Norton S. M.; Smith P. C.; Stonas W. G.; Keating C. D.; Natan M. J. Electrochemical Synthesis and Optical Readout of Striped Metal Rods with Submicron Features. J. Electroanal. Chem. 2002, 522, 95–103. [Google Scholar]
- Templeton K. E.; Scheltinga S. A.; Beersma M. F. C.; Kroes A. C. M.; Claas E. C. J. Rapid and Sensitive Method Using Multiplex Real-Time PCR for Diagnosis of Infections by Influenza A and Influenza B Viruses, Respiratory Syncytial Virus, and Parainfluenza Viruses 1, 2, 3, and 4. J. Clin. Microbiol. 2004, 42, 1564–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton I. D.; Norton S. M.; Balasingham A.; He L.; Oviso D. F.; Gupta D.; Raju P. A.; Natan M. J.; Freeman R. G. Particles for Multiplexed Analysis in Solution: Detection and Identification of Striped Metallic Particles Using Optical Microscopy. Anal. Chem. 2002, 74, 2240–2247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.