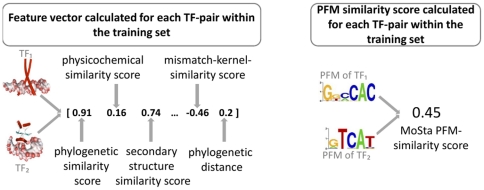
Figure 2. Training of SVR model to predict PFM similarities.
An SVR-based supervised machine learning approach is used to predict pairwise PFM similarities based on various features, derived from amino acid sequences of the DNA-binding domains of pairs of TFs. To this end, for each TF pair in the training set, a feature vector consisting of phylogenetic, physicochemical and structural domain similarity scores is computed. All pairwise PFM similarities in the training set are quantified using MoSta [36]. Next, a support vector machine is trained to predict PFM similarities based on the sequence-derived feature vectors. In machine learning, this methodology is referred to as supervised distance metric learning [33].