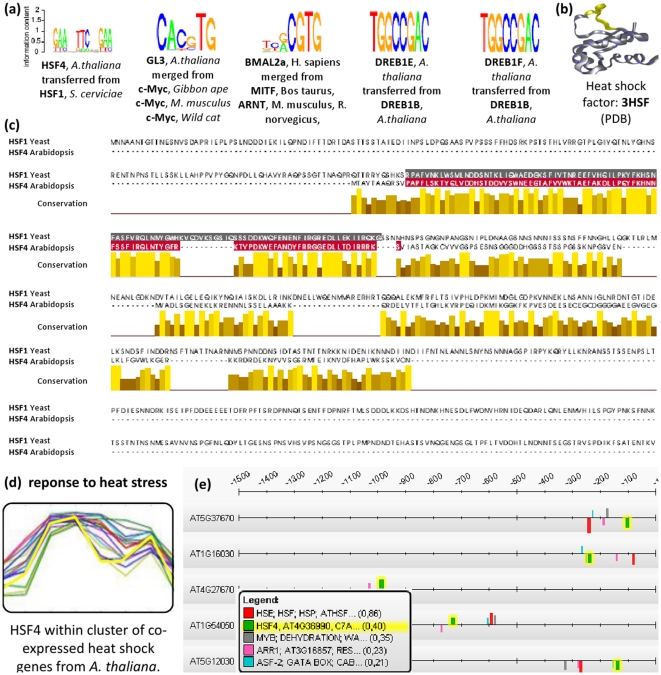
Figure 7. Examples of PFMs transferred to different TFs.
(a) Depicted are five examples of PFMs that are transferred to the query TF. These transfers merge PFMs from different species and the final PFM is depicted as sequence logo. (b) Depicts the physical structure of the DNA-binding domain of HSF4 from A. thaliana [61] that is drawn with BallView [62]. (c) For HSF4, the query and the best matching TF are aligned with JalView and their DNA-binding domains are colored [63]. This alignment contains a gap consisting of eleven amino acids in the DNA-binding domain of the HSF1 from S. cerevisiae. The amino acids that constitute the gap in the alignment are drawn yellow within the physical structure (see (b)). From this structure it can be seen that the colored amino acids do not affect the canonical helix-turn-helix structure of the HSF that is responsible for specific DNA-binding [40]. (d) Depicts A. thaliana cluster of co-expressed genes that contains HSF4 and 15 other heat shock genes, which was derived with EDISA [42]. (e) Depicts promoter scans of these genes; several matches of the predicted HSF4 PFM were detected by ModuleMaster [29].