Table 1. Comparisons with experimental parameters.
| Parameter | Simulation | Experiments | 7 Cell type | Ref. |
1Glucose uptake (kg s m m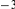 ) ) |
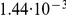
|
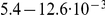
|
Rat-T1, MR1 | [37] |
1Lactate release (kg s m m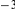 ) ) |
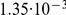
|
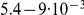
|
Rat-T1, MR1 | [37] |
2pO (mmHg) (mmHg) |
7 | 0–20 | Rat-T1 | [37] |
| 0–40 | MR1 | [37] | ||
| 20–60 | EMT6/Ro | [38] | ||
| 3pH | 6.7 | 6.6 | C6, H35 | [39] |
| 6.96–6.99 | U118-MG, HTh7 | [40] | ||
4
 pH pH |
0.77 | 0.41 | U118-MG | [40] |
0.49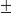 0.08 0.08 |
HTh7 | [40] | ||
5Viable cell rim thickness (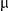 m) m) |
155 | 200 | EMT6/Ro | [38] |
142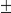 16 16 |
HTh7 | [40] | ||
310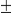 28 28 |
U118-MG | [40] | ||
198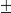 27 27 |
Col12 | [41] | ||
225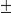 26 26 |
HT29 | [41] | ||
6Hypoxic rim thickness (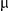 m) m) |
98 | 44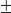 52 52 |
Col12 | [41] |
44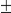 52 52 |
HT29 | [41] | ||
| Cell cycle distribution (%) | G0/G1 = 57.3 | G0/G1 = 58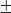 4 4 |
BMG-1 | [42] |
| S = 21.6 | S = 19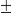 1 1 |
|||
| G2/M = 21.1 | G2/M = 23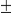 1 1 |
Metabolic and histologic parameters in spheroids of approximately 500 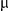 m diameter: comparison between a single, large simulation, carried out with the parameters listed in Text S1, and experimental data.
m diameter: comparison between a single, large simulation, carried out with the parameters listed in Text S1, and experimental data.
Notes:
Rate of glucose uptake or lactate release per viable spheroid volume (see [37]).
Central pO tension (experiments) or estimated in the centroid (simulations).
tension (experiments) or estimated in the centroid (simulations).
pH has been determined in the central region of the spheroids. This corresponds to a sphere of radius 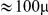 m about the centroid of the spheroid.
m about the centroid of the spheroid.
Difference between environmental pH and pH 200 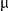 m below the spheroid surface.
m below the spheroid surface.
In our simulations the viable cell rim thickness corresponds to the distance between the spheroid surface and the inner shell where only 5% of the cells are still alive. Experimental values have been determined by histology.
These values corresponds to the radius of the necrotic core.
Cell types are as follows: Rat-T1 = T24Ha-ras-transfected Rat1 cells (Rat1 = spontaneously immortalized rat embryo fibroblasts); MR1 = myc/ T24Ha-ras-cotransfected rat embryo fibroblasts; EMT6/Ro = mouse mammary tumor cells; C6 = rat glioma cells; H35 = rat hepatoma cells; U118-MG = human glioblastoma cells; HTh7 = human tyroid carcinoma cells; Col12 = moderately differentiated human colon adenocarcinoma ; HT29 = poorly differentiated human colon adenocarcinoma; BMG-1 = human glioma cells.
