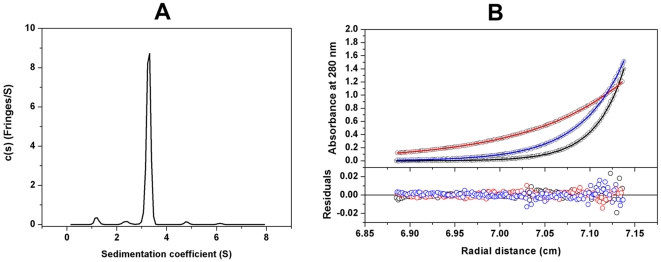
Figure 3. Analytical ultracentrifugation of the HPPK-DHPS bifunctional enzyme from Francisella tularensis.
(A) The sedimentation velocity profiles (fringe displacement) were fitted to a continuous sedimentation coefficient distribution model c(s). The experiment was conducted at a loading protein concentration of 0.69 mg/ml in at 20°C and at a rotor speed of 60,000 rpm. (B) Absorbance scans at 280 nm at equilibrium are plotted versus the distance from the axis of rotation. The protein was centrifuged at 4°C for at least 24 h at each rotor speed of 15 k (red), 22 k (blue) and 27 k (black) rpm. The solid lines represent the global nonlinear least squares best-fit of all the data sets to a monomer-dimer self-association model with a very weak KD (2.7 mM). The loading protein concentration was 20 µM and the r.m.s. deviation for this fit was 0.0037 absorbance units.