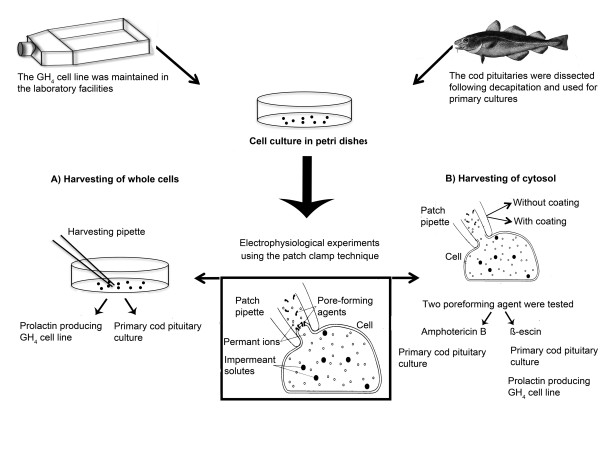
Figure 1.
Overview of the employed procedures and their validation for single-cell qPCR following electrophysiological recordings. The present study involved two different cell culture systems. Upper left, the rat pituitary GH4 cell line producing prolactin was used initially to optimize cell harvesting. Later, the cell line was also used to compare harvesting efficiency. Upper right, the main objective of this study was to set up and validate a robust assay to identify follicle-stimulating hormone (FSH)-, and luteinizing hormone (LHβ)- producing cells based on their gene expression, using primary cultures of pituitary cells from maturing cod. The cell (A)- and cytosol (B)- harvesting techniques and protocols were also adjusted to be combined with electrophysiological experiments. A) Harvesting of whole cells was performed on both the GH4 cell line and the primary cell culture from cod pituitaries. Following cell harvest, the cell was transferred to a tube containing lysis solution and then reverse transcribed into cDNA. The cDNA was precipitated using ethanol before specific gene expression was amplified using a qPCR platform. B) Harvesting of cytosol was performed using the patch pipette following giga seal formation. The patch pipette was tested both with and without silanazing the glass surface. The method was evaluated on primary cell culture from cod pituitaries. Finally, two types of cell membrane perforating agents were evaluated on the primary culture from cod pituitaries, namely amphotericin B and β-escin. The combination of using silanized patch glass and β-escin as the perforating agent was evaluated on both the GH4 cell line and primary cell culture from cod pituitaries.