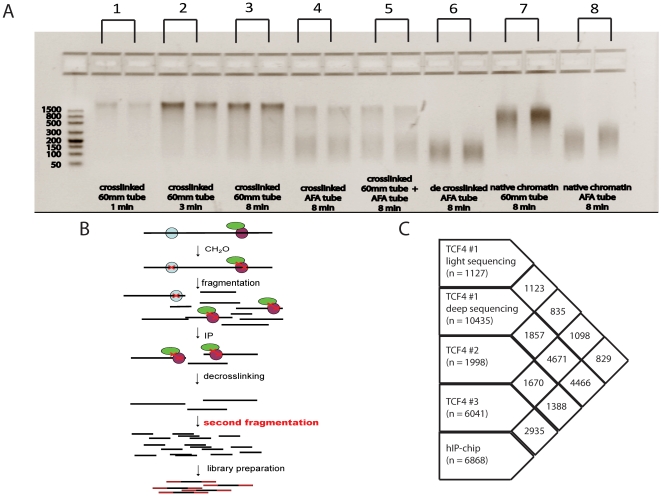
Figure 1. Double fragmentation ChIP-seq approach.
A) Comparison of different shearing methods on crosslinked, de-crosslinked and native chromatin. Samples 1–3 represent crosslinked chromatin sheared at the same power intensity with increasing shearing times in 60 mm tubes, sample 4 is crosslinked chromatin sheared using AFA tubes (Covaris), sample 5 is crosslinked chromatin sheared using 60 mm tubes and subsequently sheared in AFA tubes, sample 6 is crosslinked chromatin sheared in 60 mm tubes, de-crosslinked and subsequently sheared in AFA tubes, samples 7 and 8 are samples of native chromatin sheared using 60 mm tubes and AFA tubes, respectively. Extensive shearing of crosslinked chromatin (e.g. sample 5) still leaves a significant proportion of chromatin fragments outside the optimal range for next-generation sequencing. However, this fraction can be sheared to smaller fragments after de-crosslinking (sample 6), but not without de-crosslinking (sample 5). B) Schematic overview of the double fragmentation ChIP-seq procedure. After normal immunoprecipitation, DNA is de-crosslinked, purified and additionally sheared to concentrate all fragments in the size range that is optimal for short tag sequencers like AB/SOLiD (100–300 nt) or Illumina/Solexa (400–600 nt). C) Overlap between TCF4 ChIP-chip and ChIP-seq data. Peak sets from libraries prepared with the double shearing approach show a larger overlap with the ChIP-chip peak data.