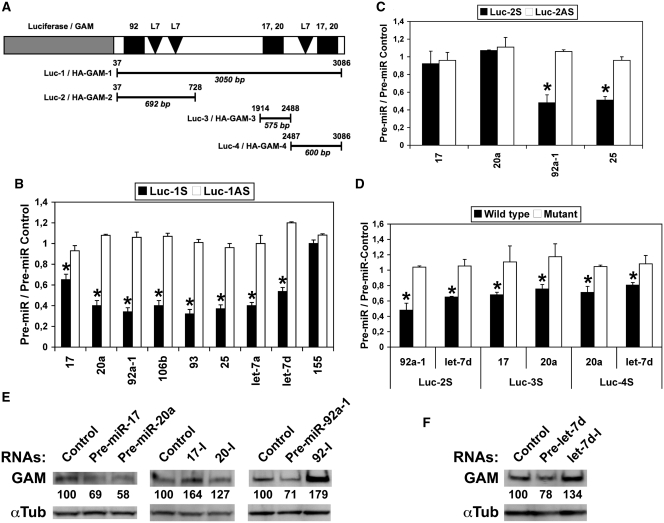
Figure 1.
GAM transcripts are direct targets of miR-17-92 and let-7 miRNAs. (A) Schematic representation of GAM 3′-UTR constructs in pGL3-Control (Luc-1, -2, -3 and -4) and pCMV-HA (HA-GAM-1, -2, -3 and -4). Conserved putative target sites for miR-17/20a (17,20), miR-92a-1 (92) and let-7 (L7) miRNAs are indicated. The positions of the 5′- and 3′-limits of the inserts with respect to GAM 3′-UTR are indicated over the lines, and their respective lengths are given in bp below the lines. (B–D) Luciferase assays were done with Luc-1 to Luc-4 as indicated, both in sense (S) and antisense (AS) orientation. Mutations in constructs used in D are given in Supplementary Tables S1 (miR-17-92 miRNA target sites) and S2 (let-7 miRNA target sites). Bars show the ratios of the Firefly luciferase (normalized to Renilla luciferase) activity measured following transfection with the indicated pre-miRNAs to that obtained following transfection with the pre-miR Control for the same construct. Values represent the mean ± SD (n = 4). (B and C) *P < 0.001, significantly different from pre-miR Control. (D) *P < 0.005, significantly different from wild type construct. (E and F) Endogenous GAM levels in HEK-293 cells transfected with either a pre-miR-Control, the indicated pre-miRNAs or antisense inhibitory RNAs to miR-17, miR-20a or miR-92a (17-I, 20-I and 92-I, respectively) were determined by western blotting.