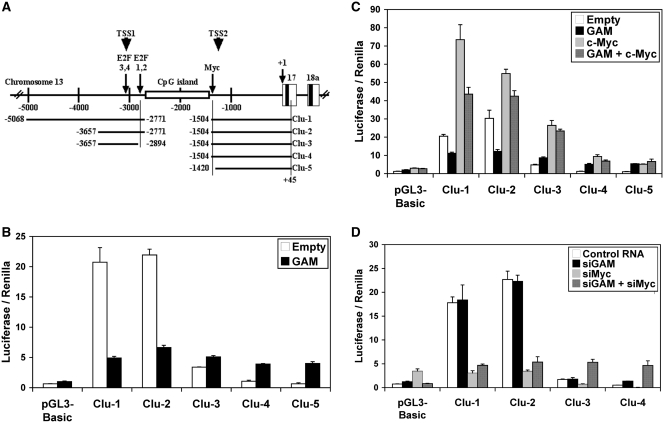
Figure 4.
GAM impairs the activation of the miR-17-92 promoter. (A) Schematic representation of the clones in pGL3-Basic containing different parts of the genomic region upstream of the miR-17-92 cluster. The first nucleotide of pre-miR-17 was arbitrarily used as nucleotide +1 for convenience. The ends of the DNA fragments used to generate the Clu-1 to Clu-5 constructs are indicated. The positions of one c-Myc-binding site (10) and of E2F-binding sites 1, 2 and 3, 4 (12) are indicated by an arrow. The approximate locations of TSS1 (transcription start site 1) (13) and TSS2 (21) are indicated by arrowheads. (B and C) Effects of GAM on the miR-17-92 promoter either alone (B) or in conjunction with c-Myc (C). MCF7 cells were transfected with the empty pGL3-Basic vector or the Clu-1 to Clu-5 constructs as indicated along with the empty pCMV-HA vector or a construct expressing GAM and/or c-Myc. The Firefly luciferase activity was measured 48 h after transfection and then normalized to the Renilla luciferase activity. Values represent the mean ± SD (n = 4). (D) Effects of siRNAs directed against GAM (siGAM) or against c-Myc transcripts (siMyc) on the miR-17-92 promoter expression either alone or in conjunction. MCF7 cells were transfected with the empty pGL3-Basic vector or the Clu-1 to Clu-4 constructs as indicated along with a control RNA, siGAM and/or siMyc. The Firefly luciferase activity was measured 48 h after transfection and then normalized to the Renilla luciferase activity. Values represent the mean ± SD (n = 4).